Human DNA BLAST Results
By now you should have the
BLAST
results back and open in another window.
We will now look at the results. The general layout, and the way the data is displayed should now be fairly familiar to you. However, we will:
- look at the different results sections
- examine the graph
- look at the sequence table
- look at, and interpret the alignment
The
BLAST
images below are taken from a search run on
Sunday October 14, 2018 at 1:47:12 pm.
The page header is always worth a glance if only to make sure you ran the right program and the right database was searched.
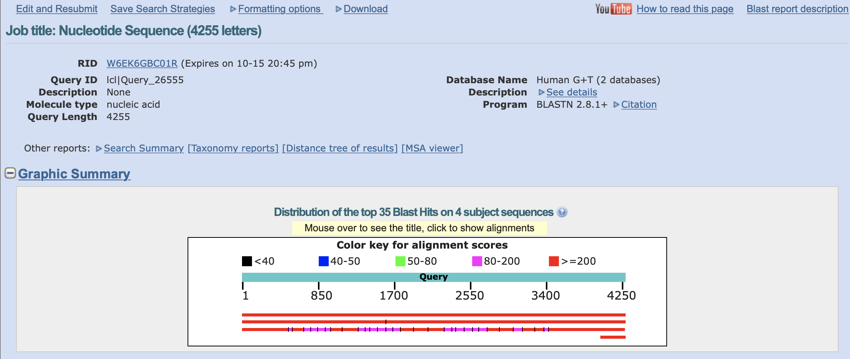
Top of the BLAST result page
The graph gives an overview of the results returned and an indication of the degree of match. Moving your mouse over the graph will show the name of the DNA.
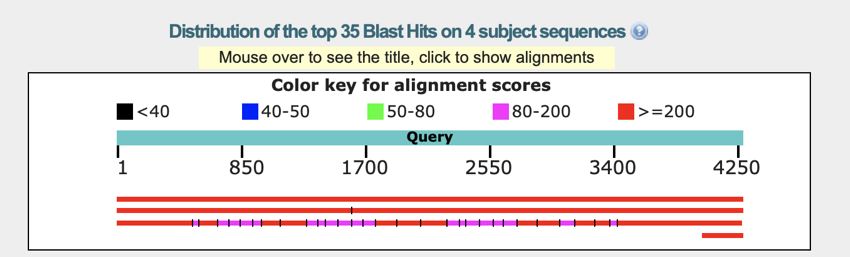
The BLAST graph - the graph shows the degree of match between your sequence and sequences in the database. In the case above you can see the matching transcript - the top line, and also the chromosomal location and intron/exon relationships.
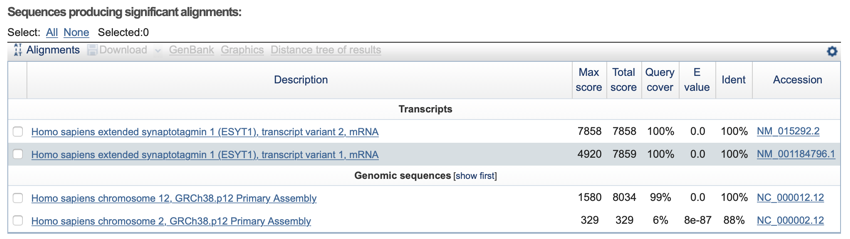
The BLAST table - the table shows the matches along with links to the full database entry and alignment. The top hit is the best match. The table is divided into transcripts and genomic data.
The
BLAST
table shows the hits ordered by best to worse.
The alignment must be examined. Just because a DNA is listed in the table, it does not mean it is a good match.
The easiest way to navigate to the alignments is by clicking on the link in the table.
- Click on the link for the top transcript alignment.
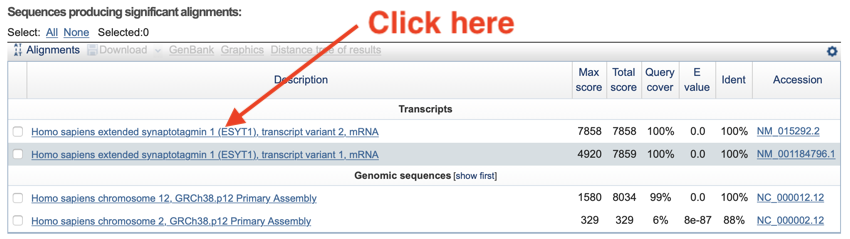
Clicking on the link - the sequence description - will take you to the alignment
The first alignment shows a very good match. This is not surprising as it is a transcript.
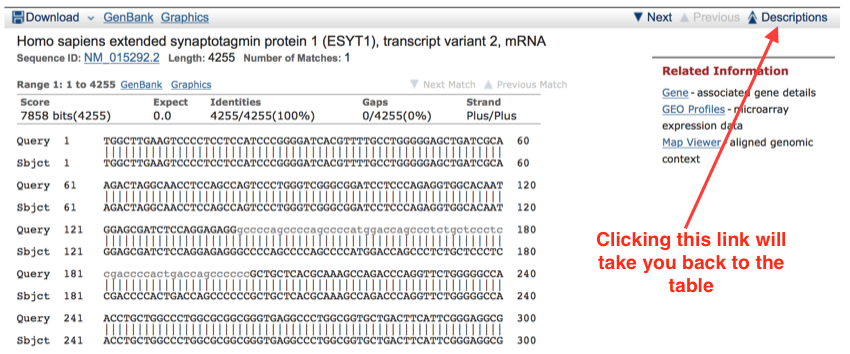
This is part of the top DNA alignment for a search run on the date shown at the top of the page. Note that this is an exact match. The identities score is 100%, and and that there are no gaps.
- Now click on the top link in the table for Genomic Sequences (Homo sapiens chromosome 12, GRCh38.p12 Primary Assembly) to take you to the first genomic match.
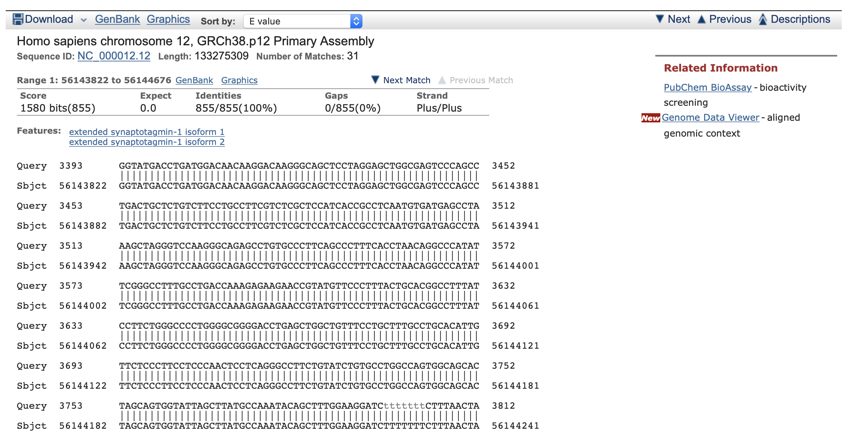
The above figure shows the first alignment in the genomic section of the results
So far we have:
- carried out a Human DNA BLAST
 search
search - looked at the returned results
- examined the sequence alignments
From these results, it is possible to look at the location and mapping to the human genome, but this is beyond the scope of this practical.
Next, we will look at searching the sequence databases with short peptide and DNA sequences.





