Running a protein BLAST
In this section, we will run a protein
BLAST
using the system at NCBI in the USA. It is also possible to run the same sort of search using a system at the European Bioinformatics Institute (
EBI
).
In this section you will be asked to:
- run a protein search for a given sequence (see below) using BLAST

- identify the article (paper) in which it was first cloned
- establish if the protein is involved in any diseases
To run a protein
BLAST
search, we need a sequence. For this search we will use (the sequence is in a straight line, so it is easy to select and copy):
MGTAGKVIKCKAAVLWEQKQPFSIEEIEVAPPKTKEVRIKILATGICRTDDHVIKGTMVSKFPVIVGHEATGIVESIGEGVTTVKPGDKVIPLFLPQVRECNACRNPDGNLCIRSDITGRGVLADGTTRFTCKGKPVHHFMNTSTFTVITVVDESSEAKIDDAAPPEKVCLIGCGFSTGYGAAVKTGKVKPGSTCVVFGLGGVGVSVIMGCKSAGASRIIGIDLNKDKFEKAMAVGATECISPKDSTKPISEVLSEMTGNNVGYTFEVIGHLETMIDALASCHMNYGTSVVVGVPPSAKMLTYDPMLLFTGRTWKGCVFGGLKSRDDVPKLVTEFLAKKFDLDQLITHVLPFKKISEGFELLNSGQSIRTVLTF
(To copy the above sequence highlight it by double click on it, and then use the copy command)
NOTE: NCBI sometimes remembers changes you have made to the default parameters. Changes are highlighted in YELLOW. Therefore, before you run a search make sure the parameters are set to the values you want (N.B. The default expect value is 10, and the default database is 'non-redundant'). To return the page to 'default' click 'Reset page' near the top of the page.
- Go to the BLAST
 site http://www.ncbi.nlm.nih.gov/blast/
site http://www.ncbi.nlm.nih.gov/blast/
- On the BLAST
 page (which should have opened in a new window) in the 'Basic BLAST
page (which should have opened in a new window) in the 'Basic BLAST ' section click the 'protein blast' link.
' section click the 'protein blast' link.
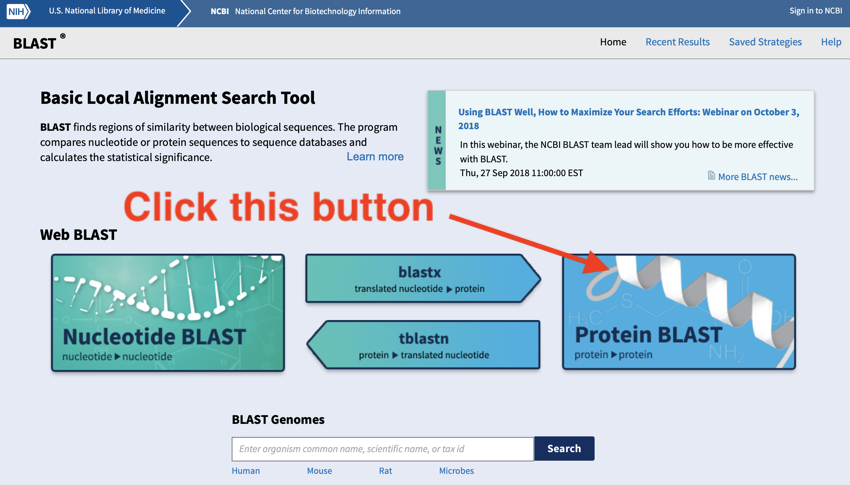
NCBI BLAST home screen - click on the protein blast button
- Paste in the data in the space provided (Don't worry about the extra spaces that are added. These will be removed during the search.)
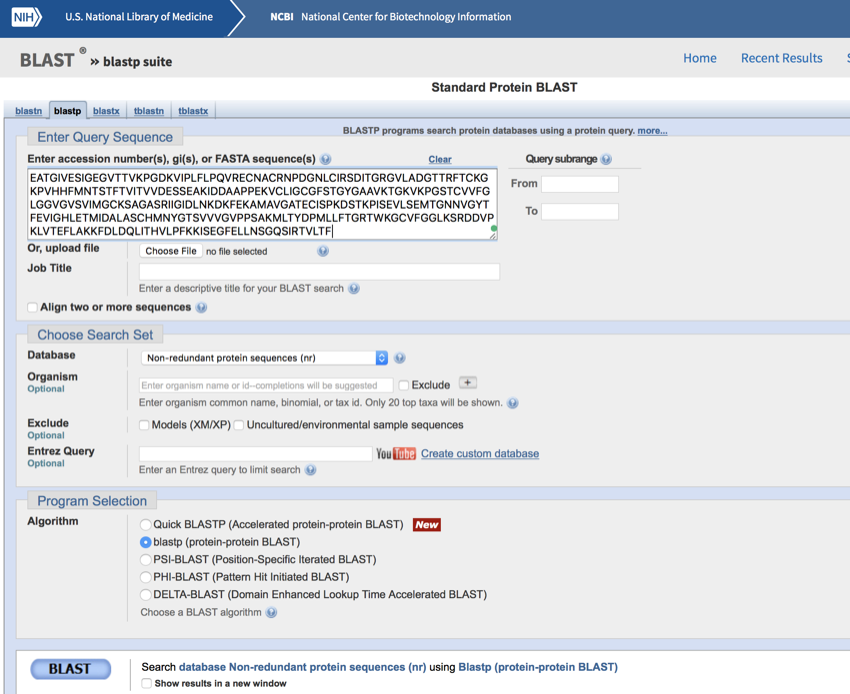
NCBI protein BLAST screen with the sequence pasted into position
- Click on the BLAST
 button.
button.
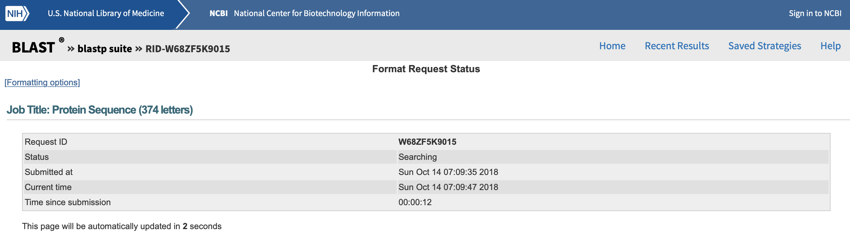
NCBI BLAST search running - search run Sunday October 14, 2018 at 12:10:25 pm
Once the result has appeared, move on to the next page.


