Protein BLAST Results
By now you should have the
BLAST
results back and open in another window.
We will now:
- look at the different results sections
- examine the graph
- look at the sequence table
- look at, and interpret the alignment
The
BLAST
images below are taken from a search run on
Sunday October 14, 2018 at 12:12:34 pm.
The page header is always worth a glance if only to make sure you ran the right program and the right database was searched.
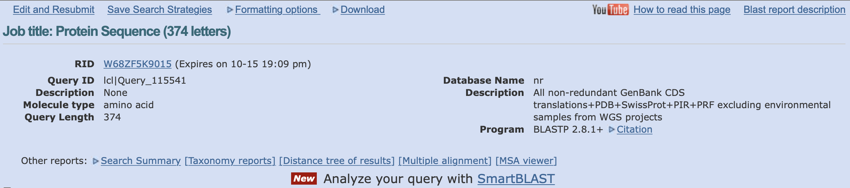
Top of the BLAST result page
The graphics section gives an overview of the putative conserved domains and the results with an indication of the degree of match.
The putative conserved domains show the regions of the protein that MAY have particular functions. This also provides an indication of how the protein may be folded in that particular region.
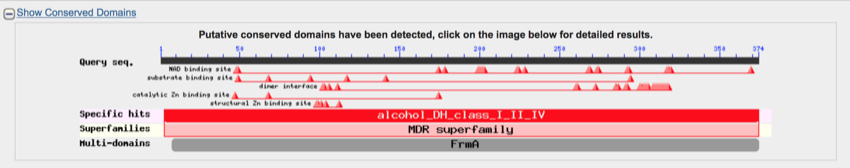
The BLAST putative domains
The small triangles show the position and occurrence of amino acids that form part of the conserved features of the domain. The large blocks indicate matches specific proteins and domains
The small triangles in the putative domains section show the position and occurrence of amino acids that form part of the conserved features of the domain. The domains can be clicked on to reveal further information (we won't be doing that in this practical).
Below the domains is the graph showing the alignments. This graph shows the degree of match (as indicated by the colour), and moving your mouse over the graph will show the name of the protein in the box (on some browsers this no longer seems to work). Clicking on one of the proteins will open a small pop-up containing additional information on the protein.
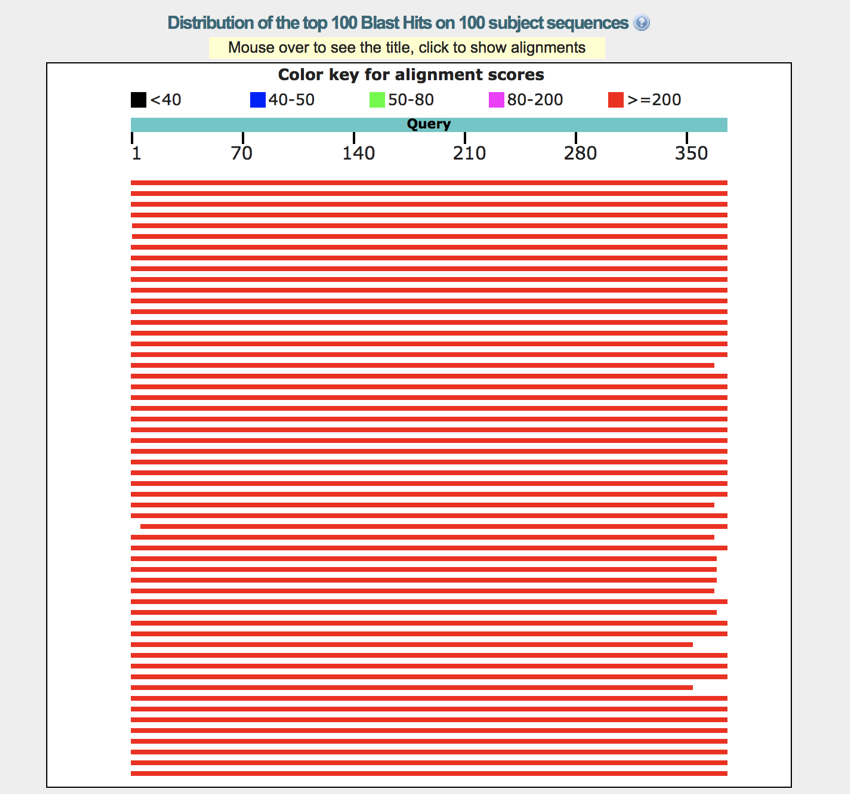
The BLAST graph - the graph shows the degree of match between your sequence and sequences in the database
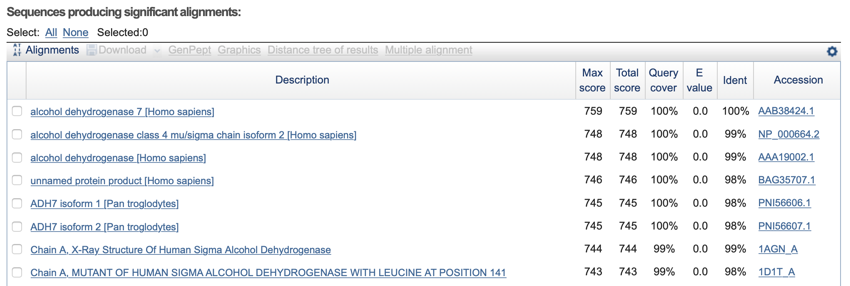
The BLAST table - the table shows the matches along with links to the full database entry and alignment. The top hit is the best match.
The
BLAST
table shows the hits ordered from best to worse (unless you click on one of the table header names such as 'Max Score' to recorder the table). The table also gives links to the alignment further down the page and the full database entry for a sequence.
The alignment must be examined. Just because a protein is listed in the table, it does not mean it is a good match. Below are two alignments.
The first alignment shows a very good (in fact exact) match.
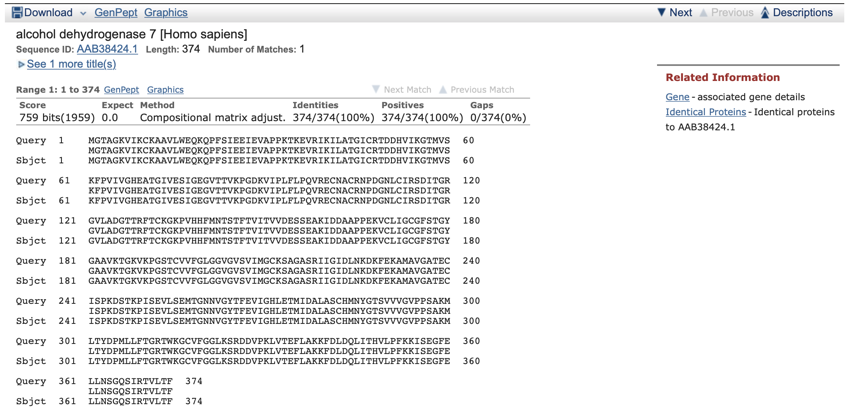
This is the top protein alignment for a search run on the date given at the top of the page. Note that this is an exact match. The identities and positives both score 100%, and that the values used to calculate the identities and positives is the same, as is the length of the protein used for the search, and for the alignments, i.e. 374 amino acids.
This alignment is not so good (this was taken from further down the list of alignments).
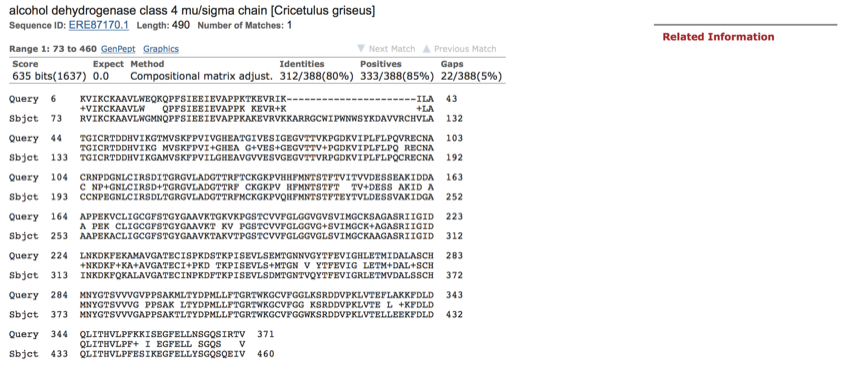
The above sequence alignment is the last one in the list and shows a poorer quality match.
This is not such as good match as although there is a full alignment across the length of the protein, the returned protein is longer (490 compared to the 374 sent), identities are 80%, and positives 85%. There is also a gap inserted.
So far we have:
- carried out a protein BLAST
 search
search - looked at the returned results
- examined the sequence alignments
We will now look at how to retrieve more information from the
BLAST
result.





