Protein Analysis - The expect value
In the previous section, you carried out a search with a sequence and got no useful information returned. No alignments, nothing. So what can we do about this?
Interestingly, before 2015 the search would have returned some very poor hits. However, the
BLAST
algorithm is constantly being improved so that now it doesn't return any results.
There are a number of reasons why this may have happened:
- The protein/DNA has not yet been sequenced
- You have used the wrong program
- You have used the wrong database
The most likely explanation is the protein hasn't been sequenced. However, one thing that can be tried is increasing the expect value.
- Go to the BLAST
 website: http://www.ncbi.nlm.nih.gov/blast/
website: http://www.ncbi.nlm.nih.gov/blast/
- Copy and Paste in the following sequence
MDAKHKNSTILGPIKAQIKAMADKVSKNALGLYHDIQVQHREYIVPISFLALILLTLANFLAGVYTFVWMLVAFKLSDVLDDVLFLRRLEKITSNVHALASNAYKQVLEESIAVESELYARFPHGEDSKQIAQIANKKKIDRWYLLDVSFRVKDKWHKKQGDMMDAKHKLGPIKAQIKAMADKVSKNALGLYHDIQVQHREYTFVWMLVAFKLSDVLDDVLFLRRLEKITSNVHALASNAYKQVLEESIAVESELYARFPHGEDSKQIAQIANKKKIDRWYLLDVSFRVKDKWHKKQGDM
- Scroll down the page and click on 'Algorithm parameters' (just below the BLAST
 button).
button). - Change the expect value to 1000
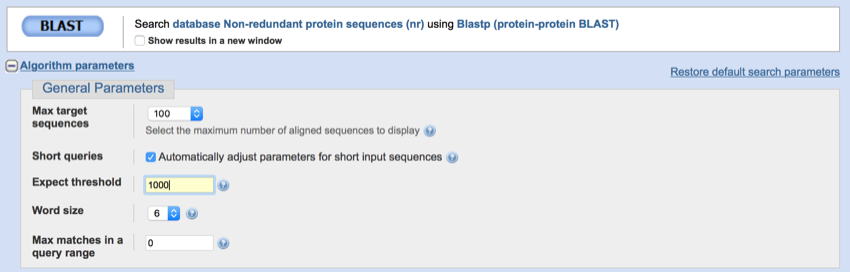
Change the expect value to 1000
- Click on the BLAST
 button
button
- Increasing the expect value has changed the number of hits returned
- Some proteins just do not match sequences in the databases
