Protein Analysis - Prosite
In the last section you carried out a
BLAST
search with a protein, and it did not return any good hits. We will now analyse the protein using
Prosite
,
Interpro
,
Pfam
and
SMART
to see if we can find any additional information from the primary amino acid sequence. First,
Prosite
.
Prosite
can analyse a sequence for the presence of a number of motifs, including:
- Post-translational modifications
- Domains
- DNA or RNA associated proteins
- Enzymes
- Oxidoreductases
- Transferases
- Hydrolases
- Lyases
- Isomerases
- Ligases
- Electron transport proteins
- Other transport proteins
- Structural proteins
- Receptors
- Cytokines and growth factors
- Hormones and active peptides
- Toxins
- Inhibitors
- Protein secretion and chaperones
- Go to the Prosite
 website at: http://prosite.expasy.org
website at: http://prosite.expasy.org
- Copy the following sequence
MDAKHKNSTILGPIKAQIKAMADKVSKNALGLYHDIQVQHREYIVPISFLALILLTLANFLAGVYTFVWMLVAFKLSDVLDDVLFLRRLEKITSNVHALASNAYKQVLEESIAVESELYARFPHGEDSKQIAQIANKKKIDRWYLLDVSFRVKDKWHKKQGDMMDAKHKLGPIKAQIKAMADKVSKNALGLYHDIQVQHREYTFVWMLVAFKLSDVLDDVLFLRRLEKITSNVHALASNAYKQVLEESIAVESELYARFPHGEDSKQIAQIANKKKIDRWYLLDVSFRVKDKWHKKQGDM
- Paste the sequence into the Prosite
 search box
search box - Set 'Exclude motifs with a high probability of occurrence from the scan' to No (remove the tick - the window should look like the figure below)
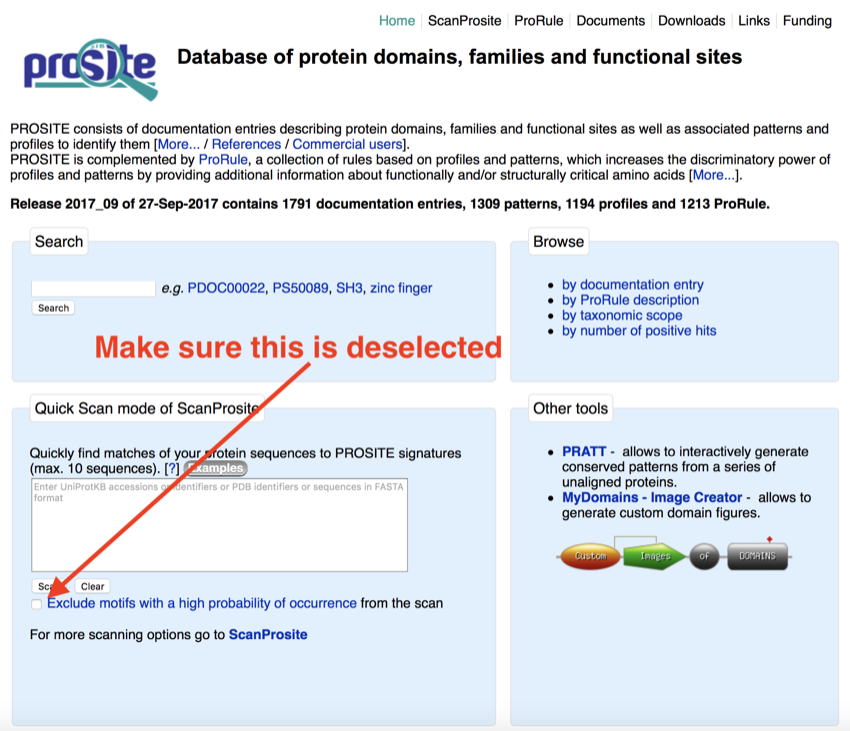
Prosite setup
- Click on 'Scan'
