Nucleosome: Difference between revisions
No edit summary |
Page still needs some references. |
||
| (7 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Nucleosome.gif|right| | [[Image:Nucleosome.gif|right|365x201px|Nucleosome.gif]]Nucleosomes are the unit of packaging within [[Eukaryotic cells|eukaryotic cells]] which allows double-stranded [[DNA|DNA]] to be packaged into the compact space inside the cell's nucleus. Nucleosomes are made up of double-stranded DNA circling [[Chromatin|chromatin]], with [[Linker histone|linker histone]] molecules joining individual nucleosomes together in a tightly compact structure. | ||
== Chromatin == | == Chromatin == | ||
| Line 10: | Line 10: | ||
*[[H4 Histone|H4]] | *[[H4 Histone|H4]] | ||
These core histones come together to form octamers that | These core histones come together to form octamers that makeup [[Chromatin|chromatin]]. H3 and H4 form a dimer and then interact with another H3/H4 dimer to form a H3/H4 tetramer. H2A and H2B also interact to form dimers. The central H3/H4 tetramer interacts with 2 flanking H2A/H2B dimers to form an octamer which forms chromatin. Approximately, DNA is wrapped around a histone twice with around 147 base pairs around it. | ||
== DNA chromatin interaction == | == DNA chromatin interaction == | ||
Double stranded DNA interacts with the molecule of chromatin by wrapping around the chromatin molecule twice | Double-stranded DNA interacts with the molecule of chromatin by wrapping around the chromatin molecule twice, forming a single [[Nucleosome|nucleosome]]. The DNA passes between individual nucleosomes, forming the 10nm fibre. The linker Histone H1 binds to the DNA between individual nucleosomes and compacts the nucleosomes into a fibre approx 30nm in diameter. | ||
This method can compact | This method can compact a genome of 2m in length (human genome) into the nucleus of a single cell. | ||
== Modification of chromatin structure == | |||
*[[Acetylation|Acetylation]] | But the action of nucleosome packaging means that transcription is inhibited as most of the DNA bound to the chromatin is inaccessible. This, therefore, requires nucleosome structure to be modulated in order to make certain strands of DNA accessible to transcription molecules such as [[RNA polymerase|RNA polymerase]]. Such methods of chromatin structure modulation include: | ||
*[[Methylation|Methylation]] | |||
*[[Ubiquitination|Ubiquitination]] | *[[Acetylation|Acetylation]] | ||
*[[Methylation|Methylation]] | |||
*[[Ubiquitination|Ubiquitination]] | |||
*[[Phosphorylation|Phosphorylation]] | *[[Phosphorylation|Phosphorylation]] | ||
These are post translation modifications which are applied to the N terminals of the | These are [[Post-translation modification|post-translation modifications]] which are applied to the N terminals of the histone molecules that make up the chromatin. These N terminals protrude from the nucleosome structure and are easily accessible. | ||
These modifications are thought to act as beacons for other proteins with [[Bromodomains|bromodomains]] to interact with the histones to make the DNA accessible to the required proteins within the cell. | |||
Latest revision as of 13:43, 17 October 2018
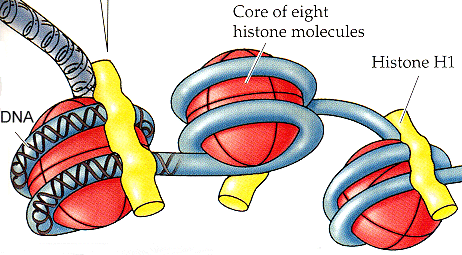
Nucleosomes are the unit of packaging within eukaryotic cells which allows double-stranded DNA to be packaged into the compact space inside the cell's nucleus. Nucleosomes are made up of double-stranded DNA circling chromatin, with linker histone molecules joining individual nucleosomes together in a tightly compact structure.
Chromatin
Chromatin is maded up of core histone molecules which are highly conserved in nature throughout most species. The main core histones are:
These core histones come together to form octamers that makeup chromatin. H3 and H4 form a dimer and then interact with another H3/H4 dimer to form a H3/H4 tetramer. H2A and H2B also interact to form dimers. The central H3/H4 tetramer interacts with 2 flanking H2A/H2B dimers to form an octamer which forms chromatin. Approximately, DNA is wrapped around a histone twice with around 147 base pairs around it.
DNA chromatin interaction
Double-stranded DNA interacts with the molecule of chromatin by wrapping around the chromatin molecule twice, forming a single nucleosome. The DNA passes between individual nucleosomes, forming the 10nm fibre. The linker Histone H1 binds to the DNA between individual nucleosomes and compacts the nucleosomes into a fibre approx 30nm in diameter.
This method can compact a genome of 2m in length (human genome) into the nucleus of a single cell.
Modification of chromatin structure
But the action of nucleosome packaging means that transcription is inhibited as most of the DNA bound to the chromatin is inaccessible. This, therefore, requires nucleosome structure to be modulated in order to make certain strands of DNA accessible to transcription molecules such as RNA polymerase. Such methods of chromatin structure modulation include:
These are post-translation modifications which are applied to the N terminals of the histone molecules that make up the chromatin. These N terminals protrude from the nucleosome structure and are easily accessible.
These modifications are thought to act as beacons for other proteins with bromodomains to interact with the histones to make the DNA accessible to the required proteins within the cell.