Watson-Crick base pairing: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
DNA consists of two types of bases namely; [[Purine|purines]] and [[Pyrimidine|pyrimidines]]. There are two types of purines: [[Adenine|adenine]] and [[Guanine|guanine]] as well as two types of Pyrimidines: [[Cytosine|cytosine]] and [[Thymine|thymine]]. In the Watson-Crick DNA base pairing model a purine always binds with a pyrimidine, however, each purine binds to one particular type of pyrimidine. | DNA consists of two types of bases namely; [[Purine|purines]] and [[Pyrimidine|pyrimidines]]. There are two types of purines: [[Adenine|adenine]] and [[Guanine|guanine]] as well as two types of Pyrimidines: [[Cytosine|cytosine]] and [[Thymine|thymine]]. In the Watson-Crick DNA base pairing model a purine always binds with a pyrimidine, however, each purine binds to one particular type of pyrimidine. | ||
Adenine (A) binds to thymine (T) whilst, guanine (G) binds to cytosine (C); although in RNA unracil (U) is substituted for thymine (T). This base pairing is referred to as complementary, hence the base pairs are called complementary [[Base pairs|base pairs]] <ref>Hartl D., Ruvolo M. (2012) Genetics; Analysis of Genes and Genomes, 8th edition, Burlington: Jones and Barlett.</ref>. The base pairs are bound by [[Hydrogen bonds|hydrogen bonds]], although the number of H-bonds differs between base pairs. G-C base pairs are bound by three (3) hydrogen bonds whilst, A-T base pairs are bound by two (2) hydrogen bonds as illustrated in the figure on the right. | Adenine (A) binds to thymine (T) whilst, guanine (G) binds to cytosine (C); although in RNA unracil (U) is substituted for thymine (T). This base pairing is referred to as complementary, hence the base pairs are called complementary [[Base pairs|base pairs]] <ref>Hartl D., Ruvolo M. (2012) Genetics; Analysis of Genes and Genomes, 8th edition, Burlington: Jones and Barlett.</ref>. The base pairs are bound by [[Hydrogen bonds|hydrogen bonds]], although the number of H-bonds differs between base pairs. G-C base pairs are bound by three (3) hydrogen bonds whilst, A-T base pairs are bound by two (2) hydrogen bonds as illustrated in the figure on the right <ref>Becker., Kleinsmith., Hardin and Bertoni (2009) The World of the Cell, Seventh Edition, San Francisco: Pearson Education.</ref>. | ||
== | <span class="Apple-style-span" style="font-size: 18px; font-weight: bold; ">Importance </span> | ||
Watson-Crick base pairing is of very great importance as it is a deciding factor in [[Semi-conservative replication|DNA Replication]] <ref name="null">Genetic Science Learning Center (2011) Build a DNA Molecule. Learn.Genetics. Available at: http://learn.genetics.utah.edu/content/begin/dna/builddna/. [Last assessed: 26/11/2011] University of Utah</ref>. It ensures that pairs form between complementary bases only. The formation of base pairs between two non-complementary bases results in [[Mutations|gene mutations]] which can be detrimental to development of an organism. | Watson-Crick base pairing is of very great importance as it is a deciding factor in [[Semi-conservative replication|DNA Replication]] <ref name="null">Genetic Science Learning Center (2011) Build a DNA Molecule. Learn.Genetics. Available at: http://learn.genetics.utah.edu/content/begin/dna/builddna/. [Last assessed: 26/11/2011] University of Utah</ref>. It ensures that pairs form between complementary bases only. The formation of base pairs between two non-complementary bases results in [[Mutations|gene mutations]] which can be detrimental to development of an organism.<span class="Apple-style-span" style="font-size: 13px; font-weight: normal; "> </span> | ||
=== Background === | === Background === | ||
In 1953, James D Watson and Francis Crick discovered the structure of DNA using X ray Crystallography. They worked out that [[DNA|DNA]] was a double helix using Rosalind Franklin's X ray diffraction pattern <ref>BBC NEWS. Science/Nature Available: http://news.bbc.co.uk/1/hi/sci/tech/2804545.stmfckLR[accessed 2 December 2011]</ref>. At first, it was thought that [[DNA|DNA]] was made up of many chemicals, which proved too difficult to analyse, but the researchers persisitence led to the discovery of complementary base pairing. | In 1953, [[James_watson|James D Watson]] and [[Francis_Crick|Francis Crick]] discovered the structure of DNA using X ray Crystallography. They worked out that [[DNA|DNA]] was a double helix using Rosalind Franklin's X ray diffraction pattern <ref>BBC NEWS. Science/Nature Available: http://news.bbc.co.uk/1/hi/sci/tech/2804545.stmfckLR[accessed 2 December 2011]</ref>. At first, it was thought that [[DNA|DNA]] was made up of many chemicals, which proved too difficult to analyse, but the researchers persisitence led to the discovery of complementary base pairing<span class="Apple-style-span" style="font-size: 13px; font-weight: normal; "> <ref>DNA tutorial Available :http://www.dnatutorial.com/BasePairing.shtml [accessed 2 December 2011]</ref>.</span> | ||
=== | === References === | ||
<references /> | <references /> | ||
Revision as of 10:04, 3 December 2011

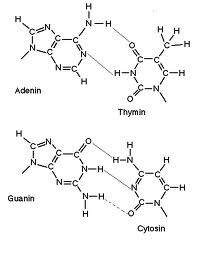
DNA consists of two types of bases namely; purines and pyrimidines. There are two types of purines: adenine and guanine as well as two types of Pyrimidines: cytosine and thymine. In the Watson-Crick DNA base pairing model a purine always binds with a pyrimidine, however, each purine binds to one particular type of pyrimidine.
Adenine (A) binds to thymine (T) whilst, guanine (G) binds to cytosine (C); although in RNA unracil (U) is substituted for thymine (T). This base pairing is referred to as complementary, hence the base pairs are called complementary base pairs [1]. The base pairs are bound by hydrogen bonds, although the number of H-bonds differs between base pairs. G-C base pairs are bound by three (3) hydrogen bonds whilst, A-T base pairs are bound by two (2) hydrogen bonds as illustrated in the figure on the right [2].
Importance
Watson-Crick base pairing is of very great importance as it is a deciding factor in DNA Replication [3]. It ensures that pairs form between complementary bases only. The formation of base pairs between two non-complementary bases results in gene mutations which can be detrimental to development of an organism.
Background
In 1953, James D Watson and Francis Crick discovered the structure of DNA using X ray Crystallography. They worked out that DNA was a double helix using Rosalind Franklin's X ray diffraction pattern [4]. At first, it was thought that DNA was made up of many chemicals, which proved too difficult to analyse, but the researchers persisitence led to the discovery of complementary base pairing [5].
References
- ↑ Hartl D., Ruvolo M. (2012) Genetics; Analysis of Genes and Genomes, 8th edition, Burlington: Jones and Barlett.
- ↑ Becker., Kleinsmith., Hardin and Bertoni (2009) The World of the Cell, Seventh Edition, San Francisco: Pearson Education.
- ↑ Genetic Science Learning Center (2011) Build a DNA Molecule. Learn.Genetics. Available at: http://learn.genetics.utah.edu/content/begin/dna/builddna/. [Last assessed: 26/11/2011] University of Utah
- ↑ BBC NEWS. Science/Nature Available: http://news.bbc.co.uk/1/hi/sci/tech/2804545.stmfckLR[accessed 2 December 2011]
- ↑ DNA tutorial Available :http://www.dnatutorial.com/BasePairing.shtml [accessed 2 December 2011]