Cytosine: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:Cytosine-chemical-structure.png|thumb]] | [[Image:Cytosine-chemical-structure.png|thumb|Cytosine-chemical-structure.png]] | ||
Cytosine (C) is one of the four bases in [[DNA|DNA]]. It is also one of the two [[Pyrimidine|pyrimidine]] bases in [[DNA|DNA]] or [[RNA|RNA]]. | Cytosine (C) is one of the four bases in [[DNA|DNA]]. It is also one of the two [[Pyrimidine|pyrimidine]] bases in [[DNA|DNA]] or [[RNA|RNA]]. | ||
It forms complementary base | It forms complementary base pairs with [[Guanine|Guanine]] (G) with three [[Hydrogen bonds|hydrogen bonds]]. This means that fragments of [[DNA|DNA]] and [[RNA|RNA]] rich in [[Guanine|Guanine]]-Cytosine (GC) pairing have higher melting points than those richer in [[Adenine|Adenine]]-[[Thymine|Thymine]] (AT) pairing, as [[Adenine|Adenine ]](A) and [[Thymine|Thymine]] (T) only bond with two [[Hydrogen bonds|hydrogen bonds]].<br> | ||
In [[Deamination mutations|deamination mutations]], Cytosine (C) loses an | In [[Deamination mutations|deamination mutations]], Cytosine (C) loses an NH<sub>2</sub>/amine group and becomes [[Uracil|Uracil]] (U), an [[RNA|RNA]] base which base pairs with [[Adenine|Adenine]] (A). The complimentary strand at this gene loci will now have an [[Adenine|Adenine]] (A) where it originally had a [[Guanine|Guanine]] (G) so the base sequence has changed. These kinds of mutations cause SNP[[Snp's|s]] (single nucleotide polymorphisms); single base changes in the sequence of a gene that may change the amino acid coded for and, therefore, the structure of the protein expressed. This simple structural change to Cytosine (C) can have drastic effects as the [[Protein|protein]] synthesised may be non-functional <ref>Hartl, DL. and Jones, EW., 2005, Genetics: Analysis of Genes and Genomes, Sixth Ed., Jones and Bartlett Publishers. pp. 616-619.</ref>. | ||
=== References: === | === References: === | ||
<references /><br> | <references /><br> | ||
Revision as of 11:07, 18 October 2012
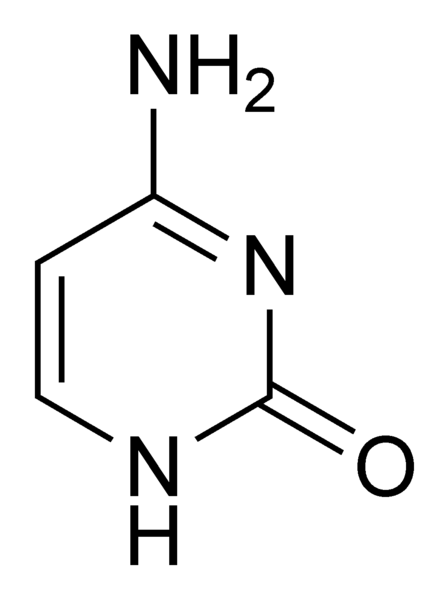
Cytosine (C) is one of the four bases in DNA. It is also one of the two pyrimidine bases in DNA or RNA.
It forms complementary base pairs with Guanine (G) with three hydrogen bonds. This means that fragments of DNA and RNA rich in Guanine-Cytosine (GC) pairing have higher melting points than those richer in Adenine-Thymine (AT) pairing, as Adenine (A) and Thymine (T) only bond with two hydrogen bonds.
In deamination mutations, Cytosine (C) loses an NH2/amine group and becomes Uracil (U), an RNA base which base pairs with Adenine (A). The complimentary strand at this gene loci will now have an Adenine (A) where it originally had a Guanine (G) so the base sequence has changed. These kinds of mutations cause SNPs (single nucleotide polymorphisms); single base changes in the sequence of a gene that may change the amino acid coded for and, therefore, the structure of the protein expressed. This simple structural change to Cytosine (C) can have drastic effects as the protein synthesised may be non-functional [1].
References:
- ↑ Hartl, DL. and Jones, EW., 2005, Genetics: Analysis of Genes and Genomes, Sixth Ed., Jones and Bartlett Publishers. pp. 616-619.