Running a DNA BLAST
You can now close all the other open windows (try not to close this one!).
In this section, we will run a DNA
BLAST
using the system at NCBI in the USA. It is also possible to run the same sort of search using a system at the European Bioinformatics Institute (
EBI
).
To run a DNA
BLAST
search we need a sequence. For this search we will use:
AGCCCTCAGGGCAGTCCCCGGCCACGGACTCCACGCGGGAGCGGGACGGGGGCTCTGGGGTCCCACCTGCCGGCCCAGGTGCGGCGAGCGAGGCCCTGGCGGTGCTGACTTCCTTCGGACGCCGCTTGTTGGTGCTGGTGCCGGTGTACCTGGCAGGGGCAGCGGGTCTTAGCGTGGGTTTCGTGCTTTTCGGCCTCGCCCTCTACCTGGGCTGGCGCCGGGTCCGCGACGGGAAAGAACGGAGCCTGAGGGCAGCGAGGCAGCTGCTGGATGACGAGGAGCGGATCACGGCAGAAACGCTTTATATGAGCCATCGGGAACTACCTGCCTGGGTCAGCTTCCCAGATGTGGAAAAGGCGGAGTGGCTGAATAAGATTGTGGTGCAGGTATGGCCCTTCCTAGGCCAATATATGGAGAAGCTTCTGGCTGAAACGGTAGCCCCAGCTGTCCGGGGAGCTAACCCCCATCTGCAAACTTTCACATTTACTCGTGTGGAGCTGGGTGAAAAGCCAGTAAGAATCATCGGAGTCAAGGTTCACCCTAGTCAGAGAAAAGATCAGATTCTACTGGACTTGAATGTCAGCTACGTGGGGGACCTACAGATTGATGTGGAAGTTAAAAAATACTTCTGCAAAGCCGGAGTCAAGGGCATGCAGCTCCACGGTGTCTTGCGGGTGATTCTTGAGCCGCTCATAGGGGACCTTCCTATCGTGGGGGCAGTGTCCATGTTCTTCATCAAACGCCCGACACTTGACATCAACTGGACAGGGATGACCAACCTGCTGGATATCCCAGGACTTAGCTCACTCTCTGACACGATGATCATGGACTCCATTGCTGCCTTCCTTGTGCTCCCTAACCGACTGTTGGTGCCCCTTGTGCCTGACCTTCAAGATGTGGCCCAGCTGCGGTCCCCACTACCCAGGGGCATTATCCGGATTCACCTGCTGGCAGCCCGAGGTCTGAGTTCCAAGGACAAGTATGTGAAAGGCCTGATTGAGGGCAAATCGGATCCCTATGCGCTTGTTCGAGTGGGCACCCAGACATTCTGCAGCCGCGTCATAGATGAGGAGCTCAACCCTCACTGGGGAGAGACGTATGAGGTGATAGTCCATGAAGTTCCAGGACAGGAGATCGAGGTGGAGGTATTTGACAAGGATCCAGATAAAGATGACTTTCTGGGAAGAATGAAGCTGGACGTAGGGAAGGTATTACAGGCTGGAGTCCTGGATAATTGGTACCCTCTACAAGGTGGGCAAGGCCAAGTTCACTTGAGGTTAGAATGGCTATCACTTCTACCAGATGCAGAAAAGCTGGATCAGGTTCTACAGTGGAATCGAGGCATCACTTCTCGGCCAGAGCCCCCGTCAGCTGCCATCCTTGTTGTCTACTTGGATCGAGCCCAGGATCTTCCTCTGAAGAAAGGCAACAAGGAACCCAATCCCATGGTGCAACTGTCAGTTCAGGATGTGACCCAGGAGAGCAAGGCTACCTACAGCACCAACTGCCCAGTGTGGGAGGAGGCTTTCCGATTCTTCCTGCAAGACCCTCGAAGCCAGGAGCTTGATGTGCAGGTGAAGGACGACTCCAGGGCCCTGACTTTAGGGGCCCTGACCCTCCCCCTTGCTCGCCTGTTGACTGCCTCTGAACTCACCCTGGACCAGTGGTTCCAGCTCAGCAGCTCAGGCCCAAACTCCAGGCTCTACATGAAACTGGTCATGCGGATCCTGTACTTAGATTCCTCAGAAATGCGCTTGCCCACTGAGCCTGGTGCCCAGGACTGGGACAGTGAGAGTCCAGAGACAGGCAGCAGTGTGGATGCCCCACCTCGGCCCTACCACACGACTCCTAACAGCCACTTTGGGACTGAGAATGTTCTTCGGATCCATGTATTAGAAGCCCAGGACCTAATTGCCAAAGACCGTTTCTTGGGGGGGCTGGTTAAGGGCAAATCAGACCCCTACGTCAAACTGAAGGTGGCAGGAAGAAGTCTCCGGACCCACGTTGTTCGGGAGGATCTCAATCCCCGCTGGAATGAGGTTTTTGAGGTGATTGTCACATCAATCCCTGGCCAAGAGCTTGACATTGAGGTCTTTGATAAGGACTTGGACAAGGATGACTTTTTGGGCAGGTATAAAGTGGGTCTCACCACAGTGCTCAACAGTGGCTTTCTTGATGAGTGGCTGACCCTGGAGGATGTCCCATCTGGGCGCCTGCACTTGCGCCTGGAGCGTCTGAGCCCCAGACCGACTGCTGCTGAGTTAGAGGAGGTGCTGCAGGTGAACAGCCTGATCCAGACCCAGAAGAGCTCAGAGCTGGCGGCAGCCCTGCTGTCTGTCTACTTGGAGCGCTCAGAAGATCTGCCGCTCCGAAAAGGAACCAAGCCTCCCAGTCCTTATGCTATTCTAACCGTGGGAGAGACATCCCATAAAACTAAGACTGTCTCCCAGACTTCAGCCCCCATCTGGGAGGAGAGTGCCTCCTTTCTCATCAGGAAGCCACATGCTGAGAGCCTGGAGCTGCAGGTTCGGGGTGAAGGGACAGGCACCTTGGGCTCCATATCCCTGCCTCTCTCTGAGCTCCTCCAGGAAGAGCAACTCTGCCTGGACCGCTGGTTTGCACTCAGCGGTCAGGGTCAGGTGCTAATGAGAGTGCAGCTAGGGATCCTGGTATCCCAGCATTCAGGAGTGGAAGCCCACAGCCACAGCTCCTCCTCTCTCAATGAAGAGCCTGAGGTTTTAGGGGACCCCACTCACACCGCCTCCCCAGTCTTAGAGGTCAGGCATCGCCTGACACATGGTGACAGTCCCTCTGAGGCTTTGATTGGGCCCCTGGGCCAGGTGAAGCTGACAGTGTGGTACCACAGTGACGAACAGAAGCTGATCAGTATTATTCATAGCTGCCGGGCCCTTCGACAAAATGGACGTGATCTCCCAGACCCCTATGTGTCAGTGTTGCTGCTCCCAGACAAGAACCGAGGCACCAAGAGGAAGACCTCACAGAAGAAGAGGACCCTCAATCCTGAGTTCAATGAGCGGTTTGAATGGGACCTGCCCCTGGATGGGACTCTTAGGAGGAAGCTCGATGTCTCTGTGAAGTCAAACTCCTCCTTCATGTCAAGAGAGCGTGAGCTTCTGGGGAAGGTGCAGCTGGATCTAGCAGAGATAGACCTTTCCCAGGGTGCAGCCCAGTGGTACGACCTGATAGATGACAGAGACAAGGGCGGTTCCTAAGAGCTGGACTGTGACAGCCTGGCACTGCTGACTTCTCGCCTCGCCTCTTGGAA
(To copy the sequence highlight it by double click on it, and then use the copy command - BTW, the reason the sequence is a big long line of letters that goes off the edge of the screen is so you can double-click on it to select it.)
And asked to:
- identify the article (paper) in which it was first cloned
- establish if the DNA is involved in any diseases
NOTE: NCBI sometimes remembers changes you have made to the default parameters. Changes are highlighted in YELLOW. Therefore, before you run a search make sure the parameters are set to the values you want (N.B. The default expect value is 10, and the default database is 'non-redundant'). To return the page to 'default' click 'Reset page' near the top of the page.
- Go to the BLAST
 site http://www.ncbi.nlm.nih.gov/blast/
site http://www.ncbi.nlm.nih.gov/blast/
- On the BLAST
 page (which should have opened in a new window) click the 'Nucleotide BLAST
page (which should have opened in a new window) click the 'Nucleotide BLAST ' button.
' button.
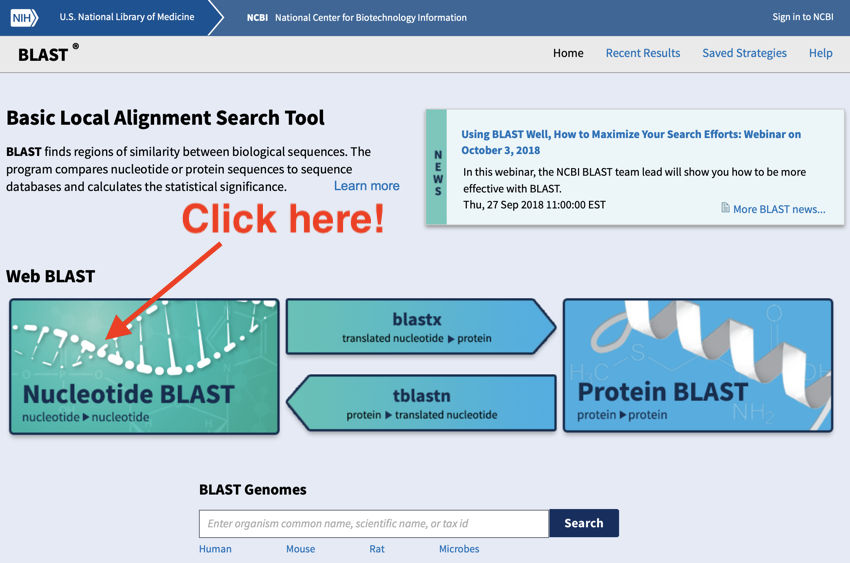
Click on the nucleotide button
- Enter (paste) the sequence into the search box
- Change the database to 'Nucleotide collection (nr/nt)' (sometimes it is set to this as a default, other times not.)
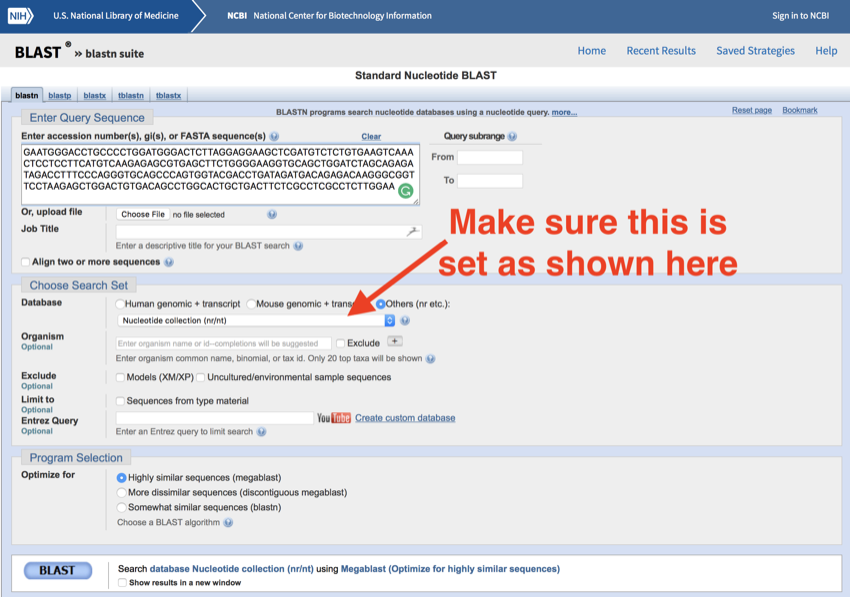
The completed nucleotide BLAST screen - make sure the database is set correctly
- Click on the BLAST
 button
button
BLAST search running - Sunday October 14, 2018 at 1:15:59 pm


