Guanosine triphosphate: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 1: | Line 1: | ||
Guanosine triphosphate (Guanosine-5'-triphosphate to be precise or also commonly abbreviated GTP for simplicity) is a high energy [[Nucleotide|nucleotide]] (not to be confused with [[Nucleoside|nucleoside]]) found in the cytoplasm or [[Polymerisation|polymerised]] to form the [[Guanine|guanine]] base. | Guanosine triphosphate (Guanosine-5'-triphosphate to be precise or also commonly abbreviated GTP for simplicity) is a high energy [[Nucleotide|nucleotide]] (not to be confused with [[Nucleoside|nucleoside]]) found in the cytoplasm or [[Polymerisation|polymerised]] to form the [[Guanine|guanine]] base. | ||
[[Image:GTP chemical structure.png|frame|right|300x200px]] | [[Image:GTP chemical structure.png|frame|right|300x200px|GTP chemical structure.png]] | ||
GTP has selective roles in the formation of [[MRNA|RNA]] strands<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>, functioning as an [[Energy carrier|energy carrier]] molecule for protein synthesis<ref>R K Murray, D A Bender, K M Botham, P J Kennelly, V W Rodwell and P A Weil. Harper's Illustrated Biochemistry. 28th Edition. Beijing, China. 2009.</ref>, a [[Coenzyme|coenzyme]], a predecessor to cGMP - a [[Secondary messenger|secondary messenger]] molecule<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref> or as an [[Effector|effector]] molecule. The last two are commonly demonstrated by [[G-protein|G-protein]] modulation<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref>. All of these are a result of it's complex three dimensional structure and the variety of different chemical groups which it comprises of. For simplicity it can therefore be thought of as a maltitude of different [[Functional group|functional groups]] that practically carry out different functions in isolation (although at times the structure will be involved e.g. when it interacts with an enzyme and another [[Substrate|substrate]]) e.g only the triphosphate is involved in releasing energy for polymerisation while only the [[Guanine|guanine base]] is involved in it's deamination. It is important to note that the list given at the start does not exhuast it's chemical interactions but is merely a demonstration of it's various capabilities. | GTP has selective roles in the formation of [[MRNA|RNA]] strands<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>, functioning as an [[Energy carrier|energy carrier]] molecule for protein synthesis<ref>R K Murray, D A Bender, K M Botham, P J Kennelly, V W Rodwell and P A Weil. Harper's Illustrated Biochemistry. 28th Edition. Beijing, China. 2009.</ref>, a [[Coenzyme|coenzyme]], a predecessor to cGMP - a [[Secondary messenger|secondary messenger]] molecule<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref> or as an [[Effector|effector]] molecule. The last two are commonly demonstrated by [[G-protein|G-protein]] modulation<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref>. All of these are a result of it's complex three dimensional structure and the variety of different chemical groups which it comprises of. For simplicity it can therefore be thought of as a maltitude of different [[Functional group|functional groups]] that practically carry out different functions in isolation (although at times the structure will be involved e.g. when it interacts with an enzyme and another [[Substrate|substrate]]) e.g only the triphosphate is involved in releasing energy for polymerisation while only the [[Guanine|guanine base]] is involved in it's deamination. It is important to note that the list given at the start does not exhuast it's chemical interactions but is merely a demonstration of it's various capabilities. | ||
| Line 11: | Line 11: | ||
=== Misconception: GTP, A Baseline Building Block === | === Misconception: GTP, A Baseline Building Block === | ||
[[Image:Xanthosine monophosphate.svg.png|frame|left|200x150px | [[Image:Xanthosine monophosphate.svg.png|frame|left|200x150px]]The [[Liver|liver]] is the principal organ which synthesises purine and pyramidine nucleotides. Purine nucleotides (GTP and ATP) are synthesized by first creating [[Inosine|inosine monophosphate]] from [[ATP|ATP]], [[Glutamine|glutamine]], [[Glycine|glycine]], [[CO2|CO2]], [[Aspartate|aspartate]] and [[Formate|formate]]<ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref>. IMP | ||
[[Image:Gout2.jpg|frame|right|200x150px | [[Image:Gout2.jpg|frame|right|200x150px]] | ||
can then be modified to yield either of the molecules. | can then be modified to yield either of the molecules<ref>R K Murray, D A Bender, K M Botham, P J Kennelly, V W Rodwell and P A Weil. Harper's Illustrated Biochemistry. 28th Edition. Beijing, China. 2009.</ref>. | ||
<span style="font-size: 13.28px;">In the case of GTP formation IMP is first converted into </span>[[XMP|XMP]]<span style="font-size: 13.28px;"> by IMP </span>[[Dehydrogenase|dehydrogenase]]<span style="font-size: 13.28px;">. The resultant chemical and structural change allows for the action of GTP synthase which rapidly converts XMP into GMP</span><ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref><span style="font-size: 13.28px;">. However, GMP is not a high energy molecule as it does not posess a triphosphate so it is then phosphorylated by nucleoside phosphate kinases to firstly yield a diphosphate and eventually a triphosphate.</span><br> | <span style="font-size: 13.28px;">In the case of GTP formation IMP is first converted into </span>[[XMP|XMP]]<span style="font-size: 13.28px;"> by IMP </span>[[Dehydrogenase|dehydrogenase]]<span style="font-size: 13.28px;">. The resultant chemical and structural change allows for the action of GTP synthase which rapidly converts XMP into GMP</span><ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref><span style="font-size: 13.28px;">. However, GMP is not a high energy molecule as it does not posess a triphosphate so it is then phosphorylated by nucleoside phosphate kinases to firstly yield a diphosphate and eventually a triphosphate.</span><br> | ||
GTP is normally catabolised into insoluble [[Uric acid|uric acid]] which can then arise in the [[Urine|urine]] as [[Sodium urate|sodium urate crystals]]<ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref>. This process is considered abnormal if it takes place in the synovial joints, resulting in uric acid which is then converted into harmful [[ | GTP is normally catabolised into insoluble [[Uric acid|uric acid]] which can then arise in the [[Urine|urine]] as [[Sodium urate|sodium urate crystals]]<ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref>. This process is considered abnormal if it takes place in the synovial joints, resulting in uric acid which is then converted into harmful [[Monosodium urate|monosodium urate]] or [[Calcium pyrophosphate dihydrate|calcium pyrophosphate dihydrate]]. The prescence of such chemicals allows for the development of<span style="font-size: 13.28px;"> </span>[[Inflammation|inflammation]]<span style="font-size: 13.28px;"> and </span>[[Arthritis|arthritis]]<ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref> and t<span style="font-size: 13.28px;">he combined symptoms lead to the classification of the disease as [[Gout|gout]]. Another example of the importance of appropriate purine nucleotide catabolism is [[Severe combined immunodeficiency disease|Severe Combined Immunodeficiency Disease]] which results in the destruction of essential [[B-cells|B]] and [[T-cells|T lymphocytes]]</span><ref>M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine</ref><span style="font-size: 13.28px;">. </span> | ||
<br> | <br> | ||
| Line 25: | Line 25: | ||
<span style="background-color: initial; font-size: 17.5296px; font-weight: bold;">One Of Many RNA Base Predecesors</span> | <span style="background-color: initial; font-size: 17.5296px; font-weight: bold;">One Of Many RNA Base Predecesors</span> | ||
RNA is | There are several differences between RNA and DNA, but the primary distinction is in the structure of the nucleobases that build each polymer. RNA nucleobases contain a 2'OH group on their [[Ribose|ribose sugar]] which allows them to be significantly more chemically reactive, catalytic and unstable. It is important to note that Guanosine triphosphate is only concerned with the production of the guanine base in RNA<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref>. The analog of GTP for the formation of a guanine base in DNA is [[deoxyguanosine triphosphate|Deoxyguanosine triphosphate]]. This triphosphate contains a [[Deoxyribose_sugar|deoxyribose sugar]] without a | ||
[[Image:RNAdiNucSyn.gif|frame|right]] | [[Image:RNAdiNucSyn.gif|frame|right|RNAdiNucSyn.gif]]<span style="font-size: 13.28px;" /> | ||
| 2'OH group which makes it significantly more stable - preventing nucleophilic attacks which would cleave the phosphodiester bonds between nucleobases. This is ideal as RNA should be disintegrated after it has been translated into a protein while DNA has to remain intact for the passage of genetic information onto the daughter cell. | ||
Guanosine triphosphate will result in the formation of a guanine base as a result of the cleavage of an [[anhydride bond|anhydride bond]] and releasing alpha and gamma phosphates as [[Free phosphate|free phosphates]]. GMP will then form a phosphodiester bond with the nearby 3'OH group containing base, and supply a 3'OH group for the next RNA base<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref>. This reaction only occurs if the opposite base to GMP is [[Cytosine|cytosine]] with which it can form [[Hydrogen_bond|hydrogen bonds]] and it's rate is increased by [[RNA_polymerase|RNA polymerase]]. RNA polymerase does not align the triphosphates. It merely catalyses the reaction that occurs after this has done independantly<ref>J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.</ref>. <span style="font-size: 13.28px;">After catalysis, the molecule is part of a polynucleotide chain and is no longer known as GTP, but as the base guanine. </span> | |||
Guanosine triphosphate will result in the formation of a guanine base as a result of cleavage of | |||
<br> | <br> | ||
| Line 43: | Line 39: | ||
The underlying mechanisms of yielding energy from anhydride bond cleavage is practically the same in all triphosphates. The high energy status of triphosphates is achieved through three distinct mechanisms. The first is due to the repulsive forces | The underlying mechanisms of yielding energy from anhydride bond cleavage is practically the same in all triphosphates. The high energy status of triphosphates is achieved through three distinct mechanisms. The first is due to the repulsive forces | ||
[[Image:Tubulin.jpg|frame|right|400x300px]] | [[Image:Tubulin.jpg|frame|right|400x300px|Tubulin.jpg]] | ||
on each of the phosphate groups. This contributes to a high instability of the bonded triphosphates and a high "desire" to achieve a lower [[Gibbs free energy|energy]] status<ref>J M Berg, J L Tymoczko, G J Gatto and R Stryer. Biochemistry. Eighth Edition. City and Country Unidentified. Freeman and Co. 2015.</ref>. The second is due to | on each of the phosphate groups. This contributes to a high instability of the bonded triphosphates and a high "desire" to achieve a lower [[Gibbs free energy|energy]] status<ref>J M Berg, J L Tymoczko, G J Gatto and R Stryer. Biochemistry. Eighth Edition. City and Country Unidentified. Freeman and Co. 2015.</ref>. The second is due to | ||
[[Image:Elongation.jpg|frame|left|350x250px]]<span style="font-size: 13.28px;" /> | [[Image:Elongation.jpg|frame|left|350x250px|Elongation.jpg]]<span style="font-size: 13.28px;" /> | ||
[[Resonance|resonance stabilisation]]. When GTP is converted into GDP the amount of | [[Resonance|resonance stabilisation]]. When GTP is converted into GDP the amount of | ||
| Line 67: | Line 63: | ||
=== A Resource for Signalling === | === A Resource for Signalling === | ||
<br> [[Image:Heterog.png|frame|left|200x200px]] GTP is not only useful due to it's chemical properties, but also due to it's intricate structure. It is able to bind and regulate the activity of different signalling pathway proteins which are classified into two different groups. The first are [[G-protein|heterotrimeric GTP binding proteins]] (also known as heterotrimeric G proteins). These utilise GTP upon the activation of the [[G-protein linked receptor|G protein coupled receptor]] in order to affect other proteins in the pathway<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. The second are [[Monomeric G-protein|monomeric GTP binding proteins]] (also known as monomeric G proteins). These respond to receptors other than G protein coupled receptors<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. A typical example cited during their explanation is the [[Ras|Ras]] protein. They are able to selectively bind GTP not only due to it's specific chemistry but also due to the [[Induced fit mechanism|induced fit phenomenon]]. It is worth noting that the bound GTP will be hydrolysed after a certain period of time and thus result in an [[Auto-inactivation|auto-inactivatio]][[Auto-inactivation|n]] of the protein<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. As a result they will thereafter contain a bound GDP molecule, but will uptake GTP upon a conformational change. GTP will be taken up after the [[Conformational change|conformational change]] due to a shift in [[Affinity|affinity]] towards the GTP molecule. | <br> [[Image:Heterog.png|frame|left|200x200px|Heterog.png]] GTP is not only useful due to it's chemical properties, but also due to it's intricate structure. It is able to bind and regulate the activity of different signalling pathway proteins which are classified into two different groups. The first are [[G-protein|heterotrimeric GTP binding proteins]] (also known as heterotrimeric G proteins). These utilise GTP upon the activation of the [[G-protein linked receptor|G protein coupled receptor]] in order to affect other proteins in the pathway<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. The second are [[Monomeric G-protein|monomeric GTP binding proteins]] (also known as monomeric G proteins). These respond to receptors other than G protein coupled receptors<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. A typical example cited during their explanation is the [[Ras|Ras]] protein. They are able to selectively bind GTP not only due to it's specific chemistry but also due to the [[Induced fit mechanism|induced fit phenomenon]]. It is worth noting that the bound GTP will be hydrolysed after a certain period of time and thus result in an [[Auto-inactivation|auto-inactivatio]][[Auto-inactivation|n]] of the protein<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. As a result they will thereafter contain a bound GDP molecule, but will uptake GTP upon a conformational change. GTP will be taken up after the [[Conformational change|conformational change]] due to a shift in [[Affinity|affinity]] towards the GTP molecule. | ||
GTP can also be used as a [[Reactant|reactant to]] produce cGMP which is a relatively common secondary signalling molecule. This catalysis is triggered by the release of NO which activates guanylate cyclase which consequently produces cGMP form GTP<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. This secondary messenger can act as an effector towards protein kinases which phosphorylate and modify the action of specific proteins. | GTP can also be used as a [[Reactant|reactant to]] produce cGMP which is a relatively common secondary signalling molecule. This catalysis is triggered by the release of NO which activates guanylate cyclase which consequently produces cGMP form GTP<ref>J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.</ref>. This secondary messenger can act as an effector towards protein kinases which phosphorylate and modify the action of specific proteins. | ||
Revision as of 09:23, 5 December 2016
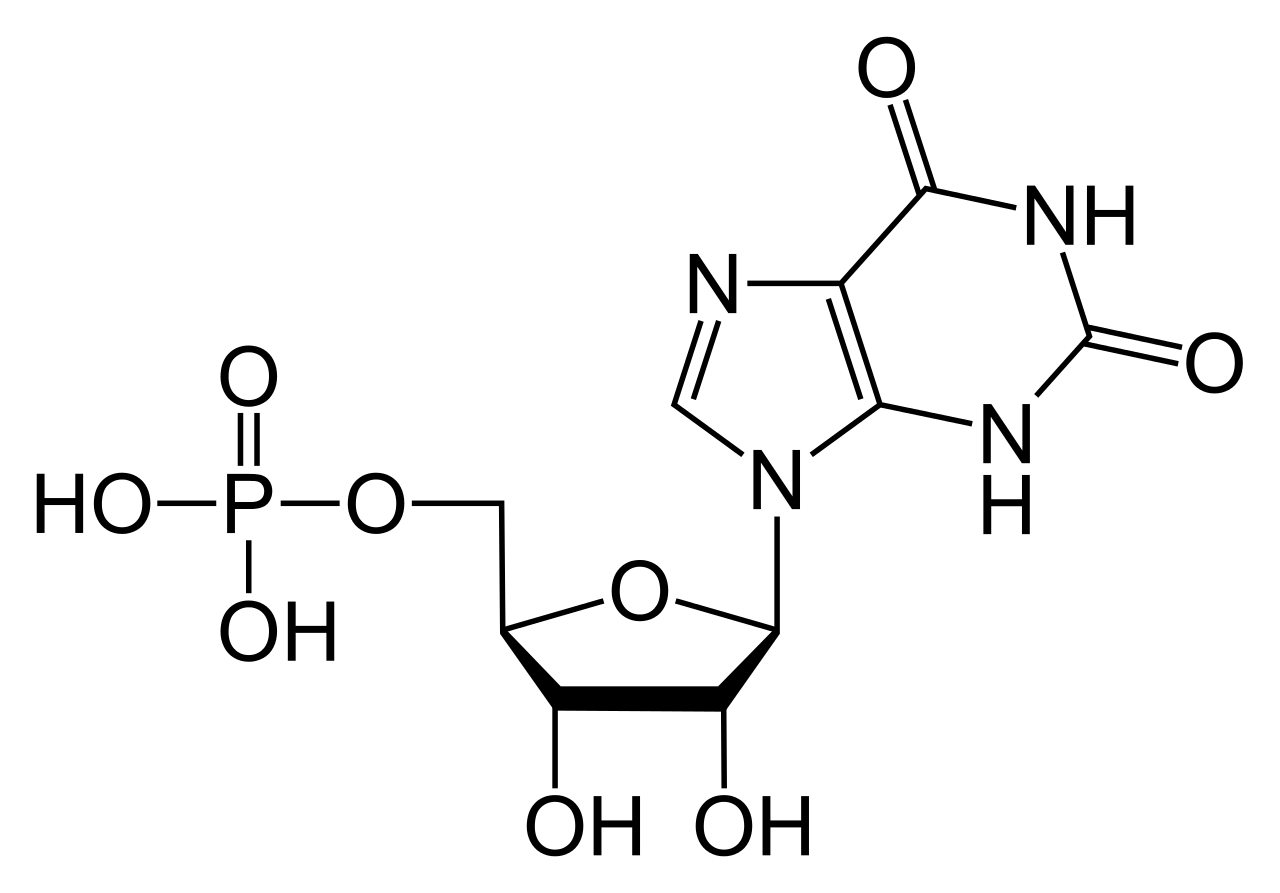
Guanosine triphosphate (Guanosine-5'-triphosphate to be precise or also commonly abbreviated GTP for simplicity) is a high energy nucleotide (not to be confused with nucleoside) found in the cytoplasm or polymerised to form the guanine base.

GTP has selective roles in the formation of RNA strands[1], functioning as an energy carrier molecule for protein synthesis[2], a coenzyme, a predecessor to cGMP - a secondary messenger molecule[3] or as an effector molecule. The last two are commonly demonstrated by G-protein modulation[4]. All of these are a result of it's complex three dimensional structure and the variety of different chemical groups which it comprises of. For simplicity it can therefore be thought of as a maltitude of different functional groups that practically carry out different functions in isolation (although at times the structure will be involved e.g. when it interacts with an enzyme and another substrate) e.g only the triphosphate is involved in releasing energy for polymerisation while only the guanine base is involved in it's deamination. It is important to note that the list given at the start does not exhuast it's chemical interactions but is merely a demonstration of it's various capabilities.
In GTP the ribose sugar is central to the three dimensional arrangement of the covalently bonded guanine and triphosphate molecules. This monosaccharide provides hydroxyl groups for condensation reactions and nucleophilic attacks[5], the latter of which is important for the destruction of RNA molecules and thus the regulation of gene expression. The guanine molecule and the triphosphate form covalent bonds at C'1 and C'5 atoms respectively, however, it is also possible for them to utilize other hydroxyl groups as long as the resultant structure does not cause clashing. From the perspective of the purine it is bonded as a result of a condensation reaction at it's 9'N, which previously had been covalently bonded to a hydrogen atom. Since guanine is a purine base, it is classified as a purine triphosphate along with adenine triphosphate (ATP)[6] and is formed through inosine monophosphate modification[7].
Misconception: GTP, A Baseline Building Block
The liver is the principal organ which synthesises purine and pyramidine nucleotides. Purine nucleotides (GTP and ATP) are synthesized by first creating inosine monophosphate from ATP, glutamine, glycine, CO2, aspartate and formate[8]. IMP
can then be modified to yield either of the molecules[9].
In the case of GTP formation IMP is first converted into XMP by IMP dehydrogenase. The resultant chemical and structural change allows for the action of GTP synthase which rapidly converts XMP into GMP[10]. However, GMP is not a high energy molecule as it does not posess a triphosphate so it is then phosphorylated by nucleoside phosphate kinases to firstly yield a diphosphate and eventually a triphosphate.
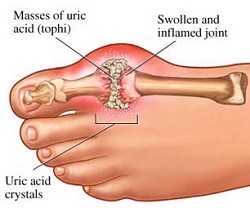
GTP is normally catabolised into insoluble uric acid which can then arise in the urine as sodium urate crystals[11]. This process is considered abnormal if it takes place in the synovial joints, resulting in uric acid which is then converted into harmful monosodium urate or calcium pyrophosphate dihydrate. The prescence of such chemicals allows for the development of inflammation and arthritis[12] and the combined symptoms lead to the classification of the disease as gout. Another example of the importance of appropriate purine nucleotide catabolism is Severe Combined Immunodeficiency Disease which results in the destruction of essential B and T lymphocytes[13].
One Of Many RNA Base Predecesors
There are several differences between RNA and DNA, but the primary distinction is in the structure of the nucleobases that build each polymer. RNA nucleobases contain a 2'OH group on their ribose sugar which allows them to be significantly more chemically reactive, catalytic and unstable. It is important to note that Guanosine triphosphate is only concerned with the production of the guanine base in RNA[14]. The analog of GTP for the formation of a guanine base in DNA is Deoxyguanosine triphosphate. This triphosphate contains a deoxyribose sugar without a
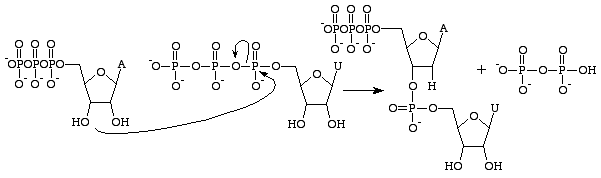
2'OH group which makes it significantly more stable - preventing nucleophilic attacks which would cleave the phosphodiester bonds between nucleobases. This is ideal as RNA should be disintegrated after it has been translated into a protein while DNA has to remain intact for the passage of genetic information onto the daughter cell.
Guanosine triphosphate will result in the formation of a guanine base as a result of the cleavage of an anhydride bond and releasing alpha and gamma phosphates as free phosphates. GMP will then form a phosphodiester bond with the nearby 3'OH group containing base, and supply a 3'OH group for the next RNA base[15]. This reaction only occurs if the opposite base to GMP is cytosine with which it can form hydrogen bonds and it's rate is increased by RNA polymerase. RNA polymerase does not align the triphosphates. It merely catalyses the reaction that occurs after this has done independantly[16]. After catalysis, the molecule is part of a polynucleotide chain and is no longer known as GTP, but as the base guanine.
GTP; A Cousin of Universal ATP
The underlying mechanisms of yielding energy from anhydride bond cleavage is practically the same in all triphosphates. The high energy status of triphosphates is achieved through three distinct mechanisms. The first is due to the repulsive forces
on each of the phosphate groups. This contributes to a high instability of the bonded triphosphates and a high "desire" to achieve a lower energy status[17]. The second is due to
resonance stabilisation. When GTP is converted into GDP the amount of
possible positions for the e
xistant electron pairs increases, lending to a greater stability[18]. This argument is also upheld with conversion into GMP. The third is due to the unfavourable entropic effect an increased amount of phosphates have on the conformation of water molecules that surround the molecule[19].
Since GTP concentration in the cytoplasm is significantly lower than that of ATP, it is used for specific functions for cell metabolic processes.
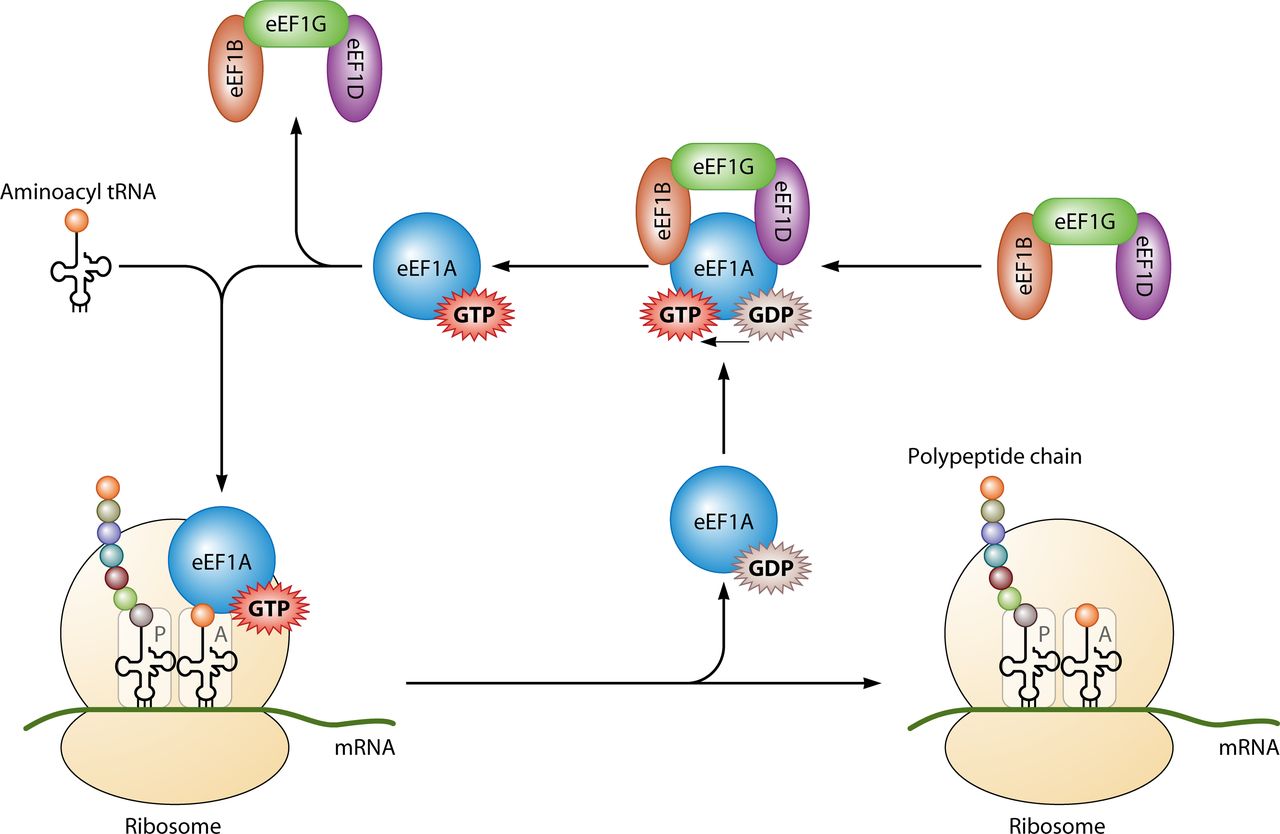
Primarily it is utilised for protein synthesis when coupled with IF2 during ribosomal initiation, Ef-Tu and
Ef-G during elongation and RF3 during termination (look left for the basic structure of the ribosomal complex) All of the GTP molecules that bind with the stated protein become hydrolysed in their distinct processes, releasing GDP and a free phosphate[20].
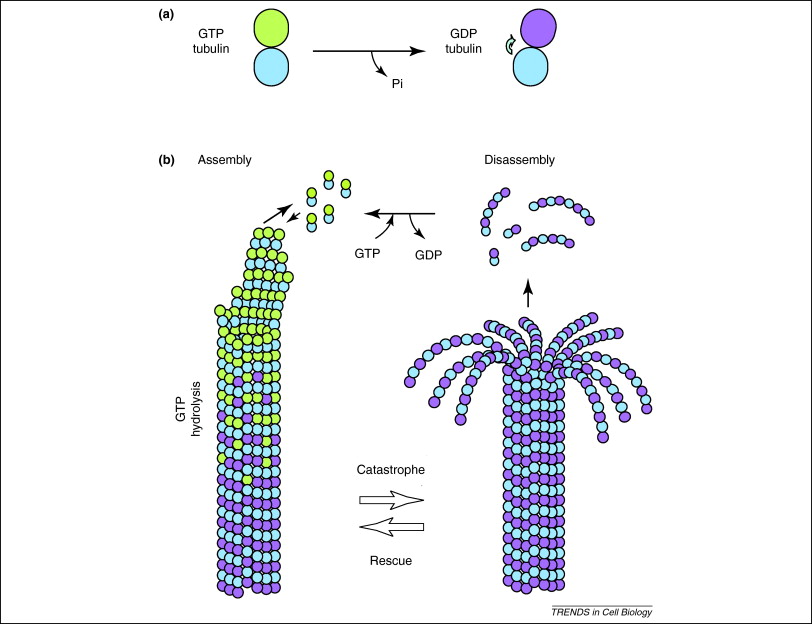
In addition, GTP is also utilised by tubulin dimers in the formation of microtubules. Tubulin dimers are composed of alpha and beta tubulin, each of which possess binding sites for GTP[21]. Since beta tubulin exists at the plus end of the filament it is always hydrolysed when another dimer is added to the lengthening polymer. The hydrolysis of GTP weakens the non-covalent interactions between tubulin dimers are results in a facilitated ability to dissolve the microtubules when necessary[22]. However, the alpha tubulin protein is not hydrolysed so it can be considered to be consistant throught the microtubule structure.
A Resource for Signalling
GTP can also be used as a reactant to produce cGMP which is a relatively common secondary signalling molecule. This catalysis is triggered by the release of NO which activates guanylate cyclase which consequently produces cGMP form GTP[26]. This secondary messenger can act as an effector towards protein kinases which phosphorylate and modify the action of specific proteins.
References
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ R K Murray, D A Bender, K M Botham, P J Kennelly, V W Rodwell and P A Weil. Harper's Illustrated Biochemistry. 28th Edition. Beijing, China. 2009.
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine
- ↑ R K Murray, D A Bender, K M Botham, P J Kennelly, V W Rodwell and P A Weil. Harper's Illustrated Biochemistry. 28th Edition. Beijing, China. 2009.
- ↑ M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine
- ↑ M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine
- ↑ M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine
- ↑ M W King. The Medical Biochemistry Page. September 2016. Cited: 11:31. 03.12.2016. Available from: http://themedicalbiochemistrypage.org/nucleotide-metabolism.php#purine
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J Dow, G Lindsay and J Morrison, Biochemistry: Molecules, Cells and the Body. 1st Edition. Wokingham, England. Addison-Wesley. 1996.
- ↑ J M Berg, J L Tymoczko, G J Gatto and R Stryer. Biochemistry. Eighth Edition. City and Country Unidentified. Freeman and Co. 2015.
- ↑ J M Berg, J L Tymoczko, G J Gatto and R Stryer. Biochemistry. Eighth Edition. City and Country Unidentified. Freeman and Co. 2015.
- ↑ J M Berg, J L Tymoczko, G J Gatto and R Stryer. Biochemistry. Eighth Edition. City and Country Unidentified. Freeman and Co. 2015.
- ↑ T E Dever and R Green. The Elongation, Termination and Recycling phases in Eukaryotes. CSHPB. July 2012. 4:7:1-16
- ↑ B Alberts et al. Molecular Biology of The Cell. 6th Edition. New York, USA. Garland Science. 2015.
- ↑ B Alberts. Molecular Biology of The Cell. 6th Edition. New York, USA. Garland Science. 2015.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.
- ↑ J Koolman and KH Roehm, Color Atlas of Biochemistry, 3rd Edition, Stuttgart, Germany. Thieme 2013.