Tutorial Using Orbital
This tutorial by
Dr. B.W. Tattershall, Chemistry, University of Newcastle upon Tyne
is in support of a course 'Atomic and Molecular Orbitals'
Contents
The Program
What is an orbital?
What is a contour plot in Orbital?
What is a Node?
Why are there Nodes?
How do Spherical Nodes in Atomic Orbitals work?
Which Quantum Numbers Count Nodes in an AO?
Getting started with an atomic orbital
Different kinds of atomic orbital node
Modelling versus Molecular Orbital Theory
What is an Antibonding Node?
What are Sigma and Pi Orbitals?
Getting started with molecular orbitals
How does Electronegativity Relate to MOs?
Polarisation
Using p orbitals
Pi molecular orbitals
MOs for HF
Sigma MOs in linear triatomic molecules
Pi MOs in linear triatomic molecules
Pi bonding to a pi-acceptor ligand
Concluding remarks
The Program
- There are two versions of the computer program needed for this tutorial,
both publicly available.
They are very similar in use
- The stand-alone program ORBITAL for Windows. This runs only in Windows-based
PCs, laptops, etc. It needs to be present on the machine and its output format
is fixed and stable
- The web application ORBITAL for Web Browsers.
This will work on either Windows or non-Windows devices.
So long as it remains available
on Newcastle University webpages, it requires no download or installation.
The output format depends on the web browser used and on window size.
Chrome, Safari or Firefox browsers have been found satisfactory, and at
least a tablet-sized window is needed for convenience of use
-
ORBITAL for Web Browsers is available by clicking the link
https://teaching.ncl.ac.uk/chemmodels/teaching/orbweb/orbital.html
- Its help is provided as a normal browser popup window
-
If you would like a copy of BWT's program ORBITAL for Windows, for your own Windows PC,
you can download it from:
-
There is a full introduction to the program, and instructions on how
to use it, compiled into its Help menu
-
It is probably best to read through the
Help from one of these sources,
before proceeding with this tutorial. You should recognise, from
the lecture course, all of the ideas discussed in the Introduction
to the Help
-
In doing these exercises, have on screen at the same time the Orbital window
and a web browser window showing this page
-
It is suggested that you write down answers to the questions, so that you
can review them as you do related exercises later in the tutorial
What is an orbital?
- An orbital is the space-dependent part of a
wavefunction which describes an electron in an atom (AO)
or in a molecule (MO). It is a mathematical
function
- It may be evaluated anywhere in space
- A surface may be drawn which joins all points at which
the wavefunction has a particular value,
e.g. ±0.04
- This may be represented in physical or graphical
rotatable models, e.g. of a
carbon p orbital in CO2
- Try switching the ball and stick model off, then pull the orbital
around, using your mouse (or finger etc. on a tablet)
- If we imagine a planar slice through the atom, and then plot contours
for regularly spaced values of the wavefunction in that plane, we can
see better how the wavefunction varies with position in space
- In the CO2 orbital window, try switching contours on,
then surfaces off, using the buttons provided. You have to wait
a little while for your computer to do the calculations
What is a contour plot in Orbital?
-
Try thinking of a contour plot of rainfall over a geographical region
-
Points of equal rainfall are joined by contour lines
-
The areas between the contour lines are coloured to make the map easier to read.
-
The Met Office uses blue for low rainfall and yellow and red for heavier rain
-
Orbital uses the same principle for displaying values of wavefunctions in a plane
through the atom(s). Warm colours from green to
dark red are used for positive values and cold colours
from cyan to deep blue are for negative values
- Grey is for less positive than green but less
negative than cyan. It therefore includes zero, so
nodes are grey, but so is all the nearly empty space
What is a Node?
- A node in an orbital is a surface where the wavefunction
changes sign
- In atomic orbitals, nodes may be either planar or
spherical
- In MOs there may be planar nodes, if they correspond
to mirror planes in the molecule, but most nodes are
other, curved shapes
Why are there Nodes?
- There is an infinity of solutions to the wave equation for an electron
in an atom or molecule, but we cut this down to a more useful set of
orbitals by choosing functions which are orthogonal to each other
- Orbitals are orthogonal if they do not overlap at all,
i.e. their overlap integral is exactly zero
- The overlap integral of two orbitals is the sum over
all space of the two functions multiplied together at each
point in space (to give the dimensions of electron
density), multiplied by a small unit of volume (to give
dimensions of amount of electron). It measures the fraction
of the electron in the overlap
- Two orbitals can have exactly zero overlap if one
sits across a node in the other, so each positive
product is cancelled out by a corresponding negative
product on the other side of the node
- Think of a 1s orbital and a 2p orbital
on the same atom. Every product of the positive
1s orbital with the positive lobe of the
2p orbital is exactly cancelled by the product at
a point symmetrically across the node, on the negative
side of the 2p orbital, so the two orbitals are orthogonal to
each other
- The same applies to two 2p orbitals: One sits across
the nodal plane of the other
- There are just two mutually orthogonal, other 2p orbitals which
can sit across the nodal plane of the first, making three in all.
Try sketching a pair of them. The two nodal planes at right angles
divide your sketch into four quarters, so products of the functions at
a point in one quarter are exactly cancelled out by products symmetrically
across one of the planes
- The nodes are in the wavefunctions in order for them
to be orthogonal to each other
- With nodes go quantum numbers, which count the nodes.
From the quantum numbers for an AO, you can
work out what nodes, planar or spherical it has,
so that you can draw it.
By looking at the contour plot of an AO, you can work out
its quantum numbers and hence give it a name, e.g.
3p
How do Spherical Nodes in Atomic Orbitals work?
- The principle is the same as for planar nodes:
a pair of orbitals differing by the presence of a spherical
node in one of them, are orthogonal because, on one
side of the node, the sum of their products,
multiplied by a small element of volume, is cancelled
out by the sum of these products on the other side of
the node
- The difference is that, unlike the planar node case,
corresponding points are not related by a plane of
symmetry of the atom, so we cannot say 'the product at
this particular point is exactly cancelled by the product at that
point'. We just accept that the constants in the
wavefunctions mean that the overlap integral calculation
gives a zero result, to within numerical error
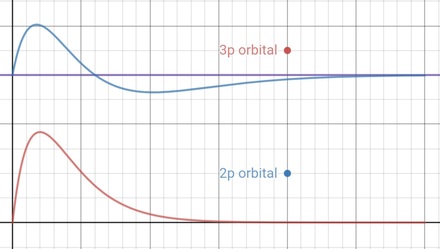
- Here are 2D plots, along the x axis, of the
2px and 3px orbitals
- The functions used come from the
Table of Functions given,
and these graphs are very similar to those often shown
in textbooks
- You may see that the spherical node in the
3px orbital comes part way along the falling
curve of the 2px orbital
- It is clear that products of the two functions to the
right of this value of x will cancel products to
the left, but not at all clear that the overall sum of
products will come out to zero
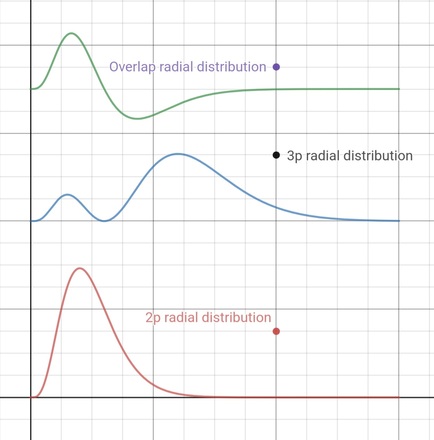
- To obtain relative amounts of electron, we square the
wavefunctions to get electron densities, then multiply
by the elements of volume. But the elements of
volume get bigger, the further from the nucleus we are:
they are proportional to r2,
or here, where we are plotting along the x axis, to
x2, so we multiply the squares of
the two functions by x2
- The resulting functions, shown here, are called
radial distribution functions
- They show, here, that most of a 3px
electron is further from the nucleus than most of
a 2px electron, but its inner lobe
penetrates the n=2 shell
- Possibly more interesting in the present context
is the top graph, which shows the product of the two
functions (which has the dimensions of electron density),
multiplied by x2 in the same way, to
give the radial distribution of overlap
- I find it easy now to believe that the total area under
this curve is zero, as required for orthogonality, which it
is to within a very small error
Which Quantum Numbers Count Nodes in an Atomic Orbital?
- The Principal Quantum Number n counts the total number
of nodes in an AO
- There are n-1 nodes altogether in an AO, not counting the one
at infinite distance which you can never see in a plot
- Thus, a 1s orbital has no visible nodes
- An n=2 orbital has one node: this is spherical
in a 2s orbital or planar in a 2p orbital
- The l Quantum Number counts the number of planar nodes
in an AO, as follows
- A l=0 orbital has no planar nodes, so is an s orbital
- A l=1 orbital has one planar node, so is a p orbital
- A l=2 orbital has two planar nodes, so is a d orbital
- If l≠0 there are 2l+1 similar orbitals in a set,
e.g. three p orbitals
- The remaining n-l-1 AO nodes, if (n-1)≠l,
are spherical nodes
Exercise 1: Getting started with an atomic orbital
-
Orbital comes up with a contour plot displayed.
Which orbital is
this? (If you have changed the plot, refresh the page)
-
How many nodes can you see, and of what kind are they
(planar or spherical)?
-
From your answers to this, write down, for this orbital,
the values of the quantum numbers n and l
respectively
- Does this correspond to the name you gave it at first?
-
The function used is given in the function box for Atom 2
-
^2 means 'raised to the power 2', i.e. squared
-
* means 'times'
-
exp means 'e to the power of '
-
What does r mean?
-
What relationships between x and y make the value of the function zero?
- Write an equation in ordinary, algebraic, form, in which
the provided function is set equal to zero
- This is the equation for the nodes:
each of its solutions describes a node
- If the expression in the first parentheses is zero,
then the whole l.h.s. is zero, so zoom in on some solutions
by writing a new equation with this expression equal to
zero. This separates x and y from r
- This is a quadratic equation, so how many solutions
does it have? Write them down
- What does the graph (y versus x) look like,
for each solution?
- Are those the nodes which you are seeing
in your Orbital window?
-
Is the function consistent with your identification of the orbital?
-
You can check your answer in the provided
Table of Functions
-
What is the effect of leaving out the constants A and B
shown in the table?
-
Start the Animation
-
What are you seeing?
-
Stop the Animation, then click Redraw Plot
-
Move the Animation Speed slider to the left, to about the 1/4 position,
or slightly less, then Start Animation again
-
Does this help you to see what is happening?
- Try to stop the animation so that the positive
and negative lobes are interchanged from the original
appearance, which corresponded to the function
- This is clearly the same orbital, at a different
instant in time, which is just as valid
- What would matter in an examination would be not
which way round you put the signs, but rather that you
did label a sketch with the signs, and that you made
sure the sign changed each time you crossed a node
(and that you marked in all the nodes and labelled
them)
-
For fun, you can set the animation speed to maximum
-
When you have had enough, stop the animation and redraw the plot
-
Use the X Range slider or 'spin wheels' or a combination of the two, to
set X Range to 5, then click Redraw Plot
-
You are plotting contours within an arbitrary window of the xy plane
-
You, the chemist, can choose what you want to plot: no particular
window is 'right'
-
This window, while it contains a very pretty plot, would not be very informative
if you did not know what you are looking at
-
Now set the X Range to 30
-
What does this tell you about where the electron, described by this function,
is to be found?
Exercise 2: Different kinds of atomic orbital node
-
Click the dropdown arrow on the function box for Atom 2
-
Starting at the top of the dropdown list and working down,
try each, in turn, of the remaining functions provided, and
identify them from the appearance of their plot
as representing particular atomic orbitals
-
Stop when you have the 3p orbital on the
screen
-
X Range should still be at 30 ; if necessary, reset it to this
-
Remember that because all the constants have been removed from these functions,
the X scale is arbitrary, and clearly not the same for this 3p function
as for the atomic orbital you looked at first. In reality, they would
be of comparable spatial extent
-
n is meant to be 3, so there should be two nodes
visible in the plot. If not,then you have selected the
wrong function
- Identify each node in the plot as spherical or planar,
and make a labelled sketch for your records
-
Slow animation may again be useful, to emphasise to you the nature of these
nodes
-
What parts of the function, shown in the Atom 2 function box, produce the
nodes which you can see? The numeric constant in
the function is arbitrary, to produce a plot which
looks like corresponding plots from real calculations
or those in textbooks
Modelling versus Molecular Orbital Theory
- Molecular Modelling is a very important part of
modern chemistry, not only for those who work in it as
a profession, but also for the very many laboratory
research chemists who do modelling calculations to predict
or inform their synthetic work or spectroscopy or whatever
- Typically, the user of a modelling package will input
or build a starting model, push the appropriate buttons,
and the package will optimise the model to a minimum energy
- This will usually involve optimising molecular geometry
at some stage
- Chemists are often most interested in predicting reaction
energetics or molecular geometry, but if the calculation
uses an electronic method ('ab initio' or
'semi empirical'), packages may offer optionally
the ability to display calculated MOs
- These are delocalised MOs which extend over the whole
molecule
- The HOMO and LUMO orbitals, and possibly MOs near to
them in energy, are often bonding in some parts and
antibonding in others. It is useful to able to
spot these features, and to see that an orbital may be
σ bonding between some atoms but π bonding
between others
- Ideas like bonding, non-bonding and antibonding, and
the terms σ bonding and π bonding, are part of
Molecular Orbital Theory, which is a concept used in
Inorganic Chemistry and Organic Chemistry to think about
bonding and molecular properties without necessarily doing
modelling calculations
- The basic principle, first current in about 1930,
is that a MO can be the sum of AOs on the constituent
atoms, each multiplied by a scaling down factor, so
that the MO represents one electron, as does each of
the contributing AOs. This is the mathematical
process of Linear Combination, so the theory is
called LCAO-MO Theory
- In this tutorial,
the equations,
for the overlap of two AOs to give a MO, are provided
on a separate web page for easy reference
- Adding together AO functions multiplied by
coefficients, and showing a contour plot
of the result, is exactly what the Orbital program does,
so you may experiment with making your own MOs
- Ab initio modelling packages also do
linear combination of atomic functions, though those
are more complex and more suited to computational
methods than the hydrogenic wave functions
considered in the MO Theory concept
- The principles are the same, and, while a good
quality ab initio modelling calculation gives
us our best idea of what 'reality' 'looks like', the
plots produced by Orbital look remarkably similar
What is an Antibonding Node?
- If the product of the coefficients of two AOs
in LCAO has the opposite sign to the overlap integral
between them, then the contribution to bonding of their
overlap in this MO is negative: it is an antibonding
contribution
- When just two AOs are overlapping, this will produce
a node in the MO, between the two atoms, perpendicular to the line
between their nuclei
- We call that an 'antibonding node'. When we
see such a node in the display of a 'real' MO, we know
that occupation of this MO would make a negative
contribution to bonding between those two atoms
- Just as nodes are present in AOs to make them
orthogonal to other AOs,
antibonding nodes are present in MOs to make them
orthogonal to corresponding bonding MOs
What are Sigma and Pi Orbitals?
- σ (sigma) and π were originally symmetry labels
which could be used for classifying MOs in linear
molecules such as H2 or CO2
- Since the middle of the 20th century, inorganic and
organic chemists have borrowed these terms to mean
MOs of similar shape to these, in molecules of lower
or no symmetry, as well as in linear ones
- We now think of a σ MO as looking like an s AO
when viewed along the bond axis between two atoms,
while a π MO looks like a p AO
- A π node is a planar node which contains the nuclei
of the two atoms concerned
- π nodes are present in MOs to make them
orthogonal to corresponding σ MOs
Exercise 3: Getting started with molecular orbitals
-
Select the 1s orbital for both Atom 1 and Atom 2
-
Change X Range to 15 and Redraw Plot
-
What kind of orbital is this?
Describe the orbital as sigma or as pi, and as bonding or as antibonding
-
Start a slow animation
-
What does this tell you about the number of nodes in this function?
Remember that a nodal surface is where a wavefunction
changes sign, not just where it gets to nearly zero
- So were you correct in your naming of the orbital?
-
Stop the animation and Redraw Plot
-
What is the sign of the overlap integral in this case?
-
What sign of the product of the coefficients is required for a bonding
combination in this case? (Remember
the equation
whose last term is the contribution to bonding
of an overlap of two AOs in an MO)
-
Change the coefficient of Atom 2 to -1
-
You may do this by clicking in front of the 1, and typing a - sign, or
you may use the 'spin wheels' if provided, or, in Orbital for Web Browsers,
by clicking the sign reverse button
-
When the coefficient is correct, click Redraw Plot
-
What is this new MO? What new feature does it
have?
-
What node(s) can you see in this function?
-
Can you see why an electron, described by this function, would be less
stable than in the AOs taken separately?
-
To investigate this, change Left Separation (i.e. between Atom 1 and Atom
2) first to 4, then to 3, doing a Redraw Plot each time, and assessing
the result. The little black dots on the plot
are the nuclei
-
Zoom in to X Range 10
How does Electronegativity Relate to MOs?
- Draw a simple energy level correlation diagram for
one orbital on one atom overlapping with one orbital
of the same type,
of different energy, on another atom,
to give a bonding and an antibonding combination
- A more electronegative atom will attract the electron
more strongly, making it more stable, so the lower
energy AO belongs to the more electronegative atom
- If we could pull the atoms apart, the bonding MO
would go up in energy as the bond got weaker, until
it was level with the more stable AO, and the
antibonding MO would come down. Each would
become identical to its respective AO
-
From this you may gather that the bonding MO looks
more like the AO of the more electronegative atom -
the classical definition of electronegativity -
but the antibonding MO is more like
the AO of the less electronegative atom
- This is of central importance in chemistry,
because most LUMOs are antibonding, so an incoming
nucleophile (or Lewis base) attacking the big end of
the LUMO will bond to the less electronegative
atom. Think of hydroxide ion attacking CO2:
It must attack the less electronegative carbon,
because that is where there is a vacant MO to accept
its electrons, not the more electronegative oxygens
Exercise 4: Polarisation
-
Still with 1s orbitals selected for Atom 1 and Atom 2, change the
coefficient of Atom 1 to 1.2 and the coefficient of Atom 2 to 0.8 (both
positive now). Redraw Plot
-
Which is the more electronegative atom, the left one (Atom 1) or the right
one (Atom 2)?
- Think of a geographical contour map: a taller
hill will usually have a bigger base
-
Change the coefficient of Atom 2 to -0.8 and Redraw Plot
-
Which is the more electronegative atom now, and how do you know?
Remember the rule explained above
Exercise 5: Using p orbitals
-
Select the 2px orbital for both Atom 1 and Atom 2
-
Change both coefficients to +1
-
Change the Left Separation to 5
-
Change the X Range to 15
-
Redraw Plot
-
What kind of orbital is this?
-
Describe the orbital as sigma or as pi, and as bonding or as antibonding
-
For what well-known diatomic molecule is this the LUMO?
- To answer this you will need to draw the
energy-level correlation diagram for the first-row
homonuclear diatomic molecules: the one without
sp mixing will do: identify the orbital
you have plotted, and count how many electrons
are needed for Aufbau, to just leave this MO empty
-
Start a slow animation
-
What does this tell you about the number of nodes in this function?
-
These nodes are of two different kinds: give names to them
(Some of these nodes were p orbital nodes in the
2px AOs used. They no longer correspond
to a mirror plane of the molecule, so they are not planar.
They do not have a universally recognised name,
but I call them p orbital nodes)
-
Is this consistent with your naming of the orbital?
-
Stop and Redraw Plot
-
What is the sign of the overlap integral in this case?
-
What sign of the product of the coefficients is required for a bonding
combination in this case? (Remember
the equation)
-
Compare your answers to those you gave to the same questions in Exercise
3
-
Change the settings so that you obtain a plot of a sigma bonding orbital
using 2p orbitals
- The antibonding node should have disappeared: notice the relative
signs of the coefficients needed to produce a bonding MO. This
is because of the sign of the overlap integral. 2px orbitals
always have their positive lobe pointing in the positive direction
along the x axis
- Compare your Orbital contour plot with the output of
a 'real'
modelling calculation for peroxide ion
- Click on the black circle marked σO(2p)O(2p) in the
Energy Level Correlation Diagram, then on the green Continue button if this
appears
Exercise 6: Pi molecular orbitals
-
Select the 2py orbital for both Atom 1 and Atom 2, and Redraw
Plot
-
Is this a bonding pi MO, or an antibonding pi MO?
-
Set the coefficients so as to make an unpolarised bonding MO, as
in the O2 molecule, and Redraw Plot
-
Why is the sign of the product of the coefficients different for a bonding
pi MO than for a bonding sigma MO using 2p orbitals?
-
Now produce a plot of the LUMO of carbon monoxide, i.e. a polarised antibonding
pi MO
-
Use coefficients -1.2 and 0.8
-
Keep Left Separation as 5, and X Range as 15
-
Which atom is carbon?
-
At which end of this MO is overlap with a transition metal pi orbital most
likely to occur?
-
Redraw the plot with Left Separation set successively to 6, 7 and 8
-
What do you notice about the shape of the lobes?
-
Separation 5 on this (arbitrary) scale is probably about right for the
equilibrium bond length of CO
- Compare your Orbital contour plot with the output of
a 'real'
modelling calculation for CO
- Click on the left-hand red circle marked π* in the
Energy Level Correlation Diagram, then on the green Continue button if this
appears
Exercise 7: MOs for HF
-
Show the sigma z MO for HF (see
Coefficients for HF) as follows
-
Select the 1s orbital for Atom 1, with a coefficient of 0.5
-
Select the 2px orbital for Atom 2, with a coefficient of -0.8
-
In program Orbital, x is used as the molecular axis, rather than the conventional
z
-
Change Left Separation to 6 (so that you can distinguish the atoms more
easily)
-
Leave X Range at 15
-
Redraw Plot
-
Why do the contours at the fluorine end of the orbital spread out further?
-
Why are opposite sign coefficients used to make this MO, when same sign
coefficients are given in the handout? (Consider the sign of the
overlap integral here, and the one given in the handout, and work out what
must be different)
-
Change the settings so as to produce a different plot of the same
orbital, using same sign coefficients
-
What do you gather from this about the sign of overlap integrals?
-
Set up a plot of the H 1s - F 2p component of the LUMO of
HF, using the coefficients given in the
table (rounded to one decimal place)
-
You should have an antibonding node as well as a p node, and the
orbital should be strongly polarised towards hydrogen
Exercise 8: Sigma MOs in linear triatomic molecules
-
Start by adding together a 2p orbital on each of the ligand atoms,
but leave out the AO on the central atom for now, as follows
-
Select None in the function list for Atom 2
-
Select 2px orbitals for both Atom 1 and Atom 3, and coefficients
of +1 for both of these
-
Set both Left Separation and Right Separation to 6
-
Set X Range to 25
-
Redraw Plot
-
At this distance apart, you see two practically unchanged p orbitals
-
With what kind of orbital on the central atom would this combination of
ligand orbital coefficients make a fully bonding MO?
-
On the basis of your answer to the last question, select the appropriate
AO for Atom 2, and set its coefficient so as to make the bonding combination
-
To obtain a reasonable representation of polarisation in CO2,
you could make the magnitude of this coefficient 0.8, and the coefficients
of the ligand orbitals 1.2
- Compare your Orbital contour plot with the output of
a 'real'
modelling calculation for carbon dioxide
- Click on the green circle marked σC(2p)O(2p) in the
Energy Level Correlation Diagram, then on the green Continue button if this
appears
-
Redraw Plot
-
This plot could also represent one 4p orbital in an octahedral first
row transition metal complex overlapping with p orbitals on trans
ligand atoms. The other two 4p orbitals do likewise, giving
three degenerate sigma p MOs
-
Now change the function for the central Atom 2 to a 1s orbital (here
used as an approximation to a 2s orbital in CO2)
-
To make the bonding MO you will now have to change the sign of one of the
ligand orbital coefficients
-
Which one needs to be changed, Atom 1 or Atom 3, to make the bonding
combination?
-
Try it, and Redraw Plot
-
From this part of the exercise, you can see that different sign combinations,
for the coefficients of the ligand orbitals, are needed for the sigma s
MO than for the sigma p MO
-
This means that s and p AOs on the central atom of a linear
triatomic molecule give separate MOs: they do not mix
-
The same applies to octahedral complexes, or to tetrahedral or trigonal
planar molecules, for the same sort of reason
-
Only in less symmetrical molecules, like bent triatomic or trigonal pyramidal,
or on the ligand atoms of molecules, do s and p AOs mix,
when they produce the MOs which contain lone pairs
-
The other important point to gather from this exercise is that the sigma
MOs (not just pi MOs as in Hueckel Theory) extend over all the atoms
-
Usually, in ordinary MO theory, there are no localised MOs: they
are all delocalised
Exercise 9: Pi MOs in linear triatomic molecules
-
Use 2py orbitals on all three atoms to create a contour plot of
an antibonding pi LUMO of CO2
-
Use coefficients of 0.8 and 1.2
-
You decide which way round these should be, and what sign they should have
-
Left and Right Separations of 5 and X Range 20 give a 'realistic'-looking
plot
-
You should see a pi node and two antibonding nodes, and it should
be apparent that a Lewis base would attack at the carbon atom, at right
angles to the molecular axis
-
If any of these features are wrong, you have erred. Revise what you
have done and correct the error
-
Make a MO setting the Atom 2 function to None, using 2py orbitals
on both atoms 1 and 3, and coefficients of +1 for Atom 1 and -1 for Atom
3. Leave both separations as 5
-
This a non-bonding pi orbital
-
It is orthogonal to the s or any of the p AOs on the central
carbon atom (demonstrate this to your own satisfaction)
-
This orbital holds one of the lone pairs on oxygen in CO2, i.e.
it is one of the HOMOs
-
Notice that this is one MO, delocalised over both oxygen atoms:
each electron of a lone pair described by this orbital will be partly on
each of the two oxygens, but not on the carbon
Exercise 10: Pi bonding to a pi-acceptor ligand
-
Imagine that Atom 1 is a transition metal in an octahedral carbonyl complex,
Atom 2 is carbon, and Atom 3 is oxygen
-
Set up a contour plot of this part of the pi system as follows
-
Select the 3dxy orbital for Atom 1, and 2py orbitals for
Atoms 2 and 3
-
Make the coefficients 1, 1.4, and -1.2 respectively, to take account of
polarisation and to create the antibonding node between C and O
-
Set Left Separation to 6, Right Separation to 4, to take account of the
different atomic radii
-
Set X Range to 23
-
Redraw Plot
-
In reality, the left lobes of this MO would carry on to another OC group
Concluding remarks
If you have succeeded in working through all of this tutorial, you should
now have a good knowledge of contour plots, of atomic wavefunctions, of
nodes in AOs and MOs, of bonding, non-bonding and antibonding sigma or
pi MOs, and of matching the signs of coefficients to the signs of overlap
integrals. You should understand polarisation and delocalisation.
You still need to cover radial distribution of AOs and energy level correlation
diagrams and orbital occupancy, none of which are dealt with here.
I hope you have enjoyed using my program Orbital and working through this
tutorial with it, as much as I have enjoyed creating them.
Bruce Tattershall, Chemistry,
University of Newcastle



