Hello DAVID!
DAVID - Database for Annotation, Visualization and Integrated Discovery (DAVID) It a rather impressive set of tools for analysing expression data.
DAVID will take the list of IDs that you generated in the last section. Remember, these IDs represent differences between gene expression in a healthy and diabetic pancreatic islet cells.
We are now going to start using DAVID.
- Go to https://david.ncifcrf.gov
 .
.
Welcome to DAVID
Don't get freaked out by the page; I'll walk you through it.
- Click on 'Start Anaylsis'.
Click on 'Start Anaylsis'
- Next, click on 'Upload'.
Click on 'Upload'
- In 'Step 1' paste in your ID list in the box provided (click on the box and then press 'Control V').
- In 'Step 2' select 'AFFYMETRIX_3PRIME_IVT_ID'(it should already be set to this).
- In 'Step 3' select 'Gene List'
Your final setup should look like the image below.
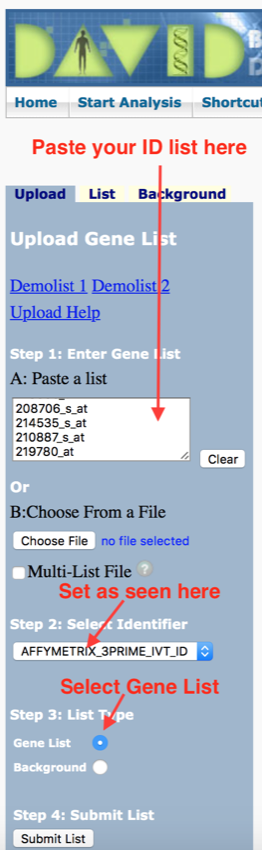
Final setup of DAVID
- Click 'Submit List'.
A new page will load...
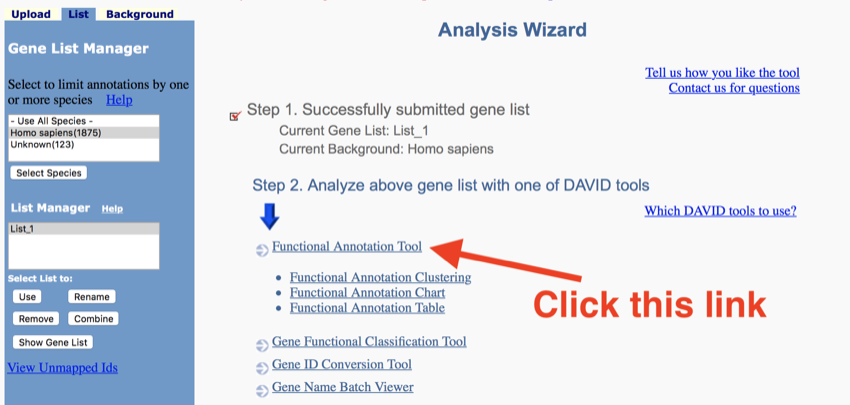
The next page of DAVID
- Click 'Functional Annotation Tool'.
You should now be on a page that looks like this...
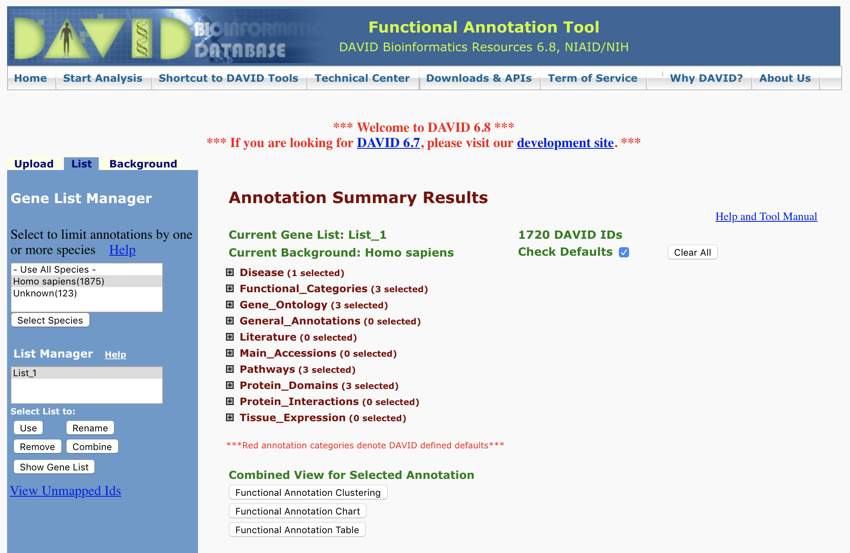
DAVID - the result - Monday November 12, 2018 at 7:06:47 pm
So what just happened?
At first glance, not a lot. However, what DAVID has done is set you up with more data analysis than you could possibly imagine.
So, now it's time to explore...
We started with a list of IDs that we have passed to DAVID, which has run a series of analyses for us.
In the next section, we are going to explore the findings.





