STRING - more than one protein
In this section we are going to look at how we can use STRING to find connections between three proteins:
- INSR
- IRS1
- AKT1
Now, I know that these three proteins are involved in insulin signalling and that INSR (the insulin receptor) and IRS1 (insulin receptor substrate 1) directly interact and that between IRS1 and AKT1 that are a number of other proteins.
So, let's see what we can find...
We will now return to the STRING site and input the three proteins.
- Go to STRING - https://string-db.org

- Click on 'Multiple proteins' in the menu on the left.
- In the box provided enter INSR, IRS1, AKT1 on sperate lines (i.e. hit the return key after each protein name) - your page should look like the image below.
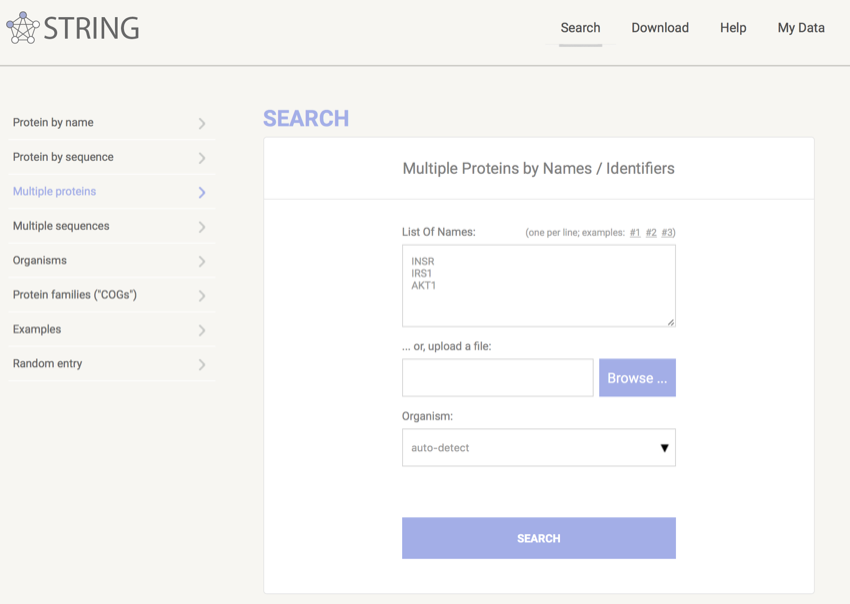
Multi-protein search in STRING
- Click on the "SEARCH" button.
You should now be looking at a list of organisms. Make sure that human is selected.
- Click on "CONTINUE"
You should now have a page open that lists the three proteins INSR, IRS1, and AKT1.
- Make sure that the correct protein is selected in each section.
- Click on "CONTINUE"
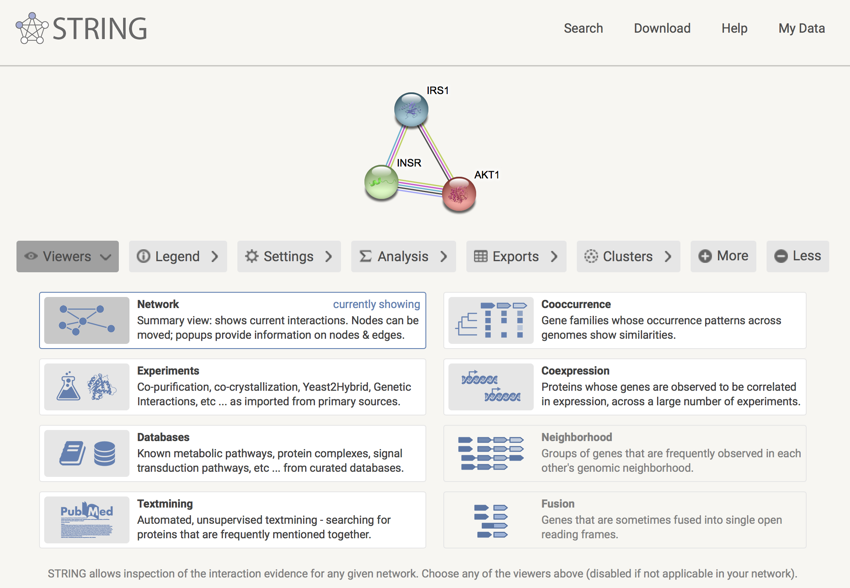
The INSR, IRS1, and AKT1 protein information network
Remember: This is a network of information. It is not a network of protein-protein interactions. In fact, the figure below shows the network of interactions with the three proteins surrounded by green-dotted boxes. As you can see, INRS and IRS1 do directly interact, but between IRS1 and AKT (also known as PKB), there is PI3K and PDK.
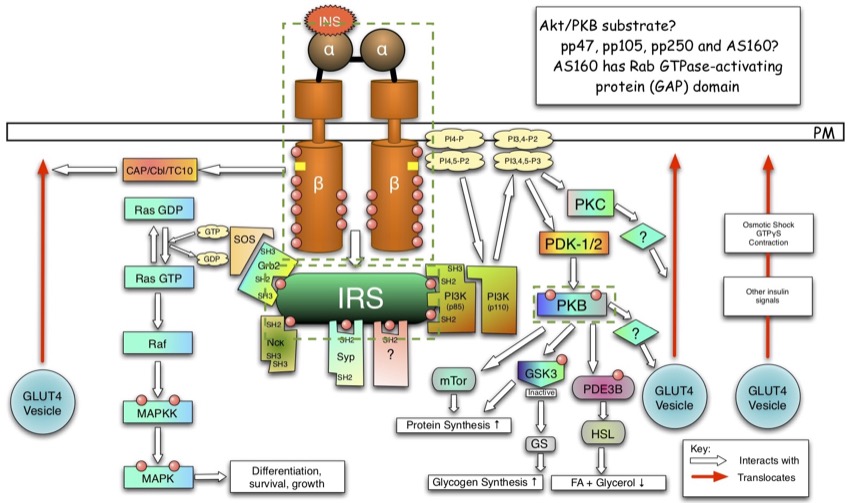
The insulin signalling pathway
- Click on the lines between INSR and IRS.
You should now see some information on INSR and IRS. Take some time to explore the links and the associated data.
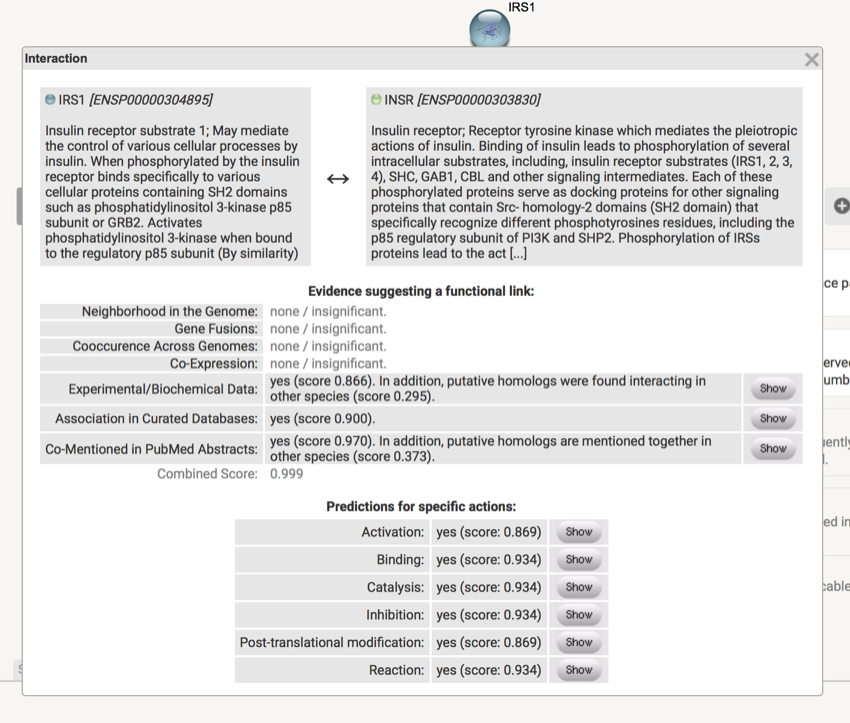
Data network associated with INSR and IRS1
- Click on the lines between IRS and AKT.
Again, take some time to explore the links and associated data.
- Click on the "Analysis" button and examine the data.
You should have discovered from the associated data that the three proteins are connected and that they are all involved in insulin signalling.
Once you have finished exploring the data return to STRING -
https://string-db.org
and repeat the above process, but this time using the following proteins:
- INSR
- IRS1
- AKT1
- GFAP
If you have entered things correctly then the result should look like this:
What do you think about the 'information' network? Does anything strike you as odd?
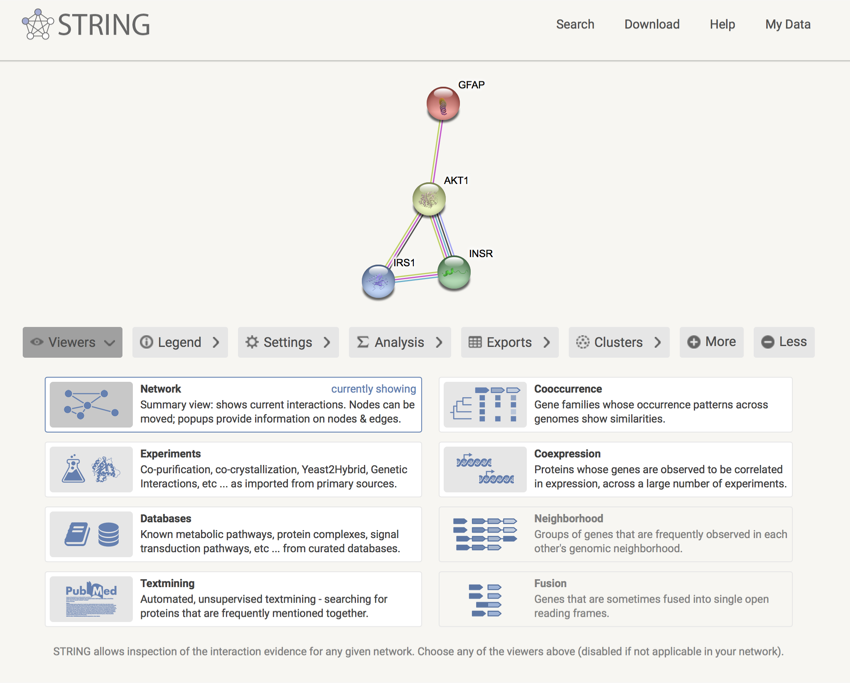
INSR, IRS1, AKT1 and GFAP information network
Take some time to explore the data associated with the information network. Specifically, don't forget to:
- Look at the Analysis data
- Explore the analysis data and look at the 'count in gene set'
- Click on the link between GFAP and AKT1
- Expand the network by clicking on 'More'
What do you think? Is GFAP a member of the INSR, IRS1, and AKT1 'club'?
If you look at the data, it is hard to see a link between INSR, IRS1 and AKT1, and GFAP. GFAP seems to stick out from the network. It only links to AKT1. There is a possible link with 'GO:0008284 positive regulation of cell proliferation', but it is not a great link. If you go 'More' and expand the network, then GFAP looks even more isolated. Looking at the 'Co-Mentioned in
PubMed
Abstracts' between GFAP and AKT1 doesn't produce any evidence of interaction (compare it to the
PubMed
data for the other proteins).
Hence, I would say that GFAP is an outlier of the dataset. And to do the above without using STRING would take many many hours of
PubMed
searches and trawling through all the different databases.
In this section, we have seen how STRING can be used to look at the information network between a number of proteins.
STRING is very useful for quickly finding information and connections between proteins and will be a very useful tool for exploring connections between any proteins of interest you discover in the assessment.




