Phylogenetics
Phylogenetics is the study of evolutionary history, achieved by analysing the molecular sequences of individual organisms[1]. It tells us how different organisms are related to each other by their evolutionary history and thus, how they have descended from one another; this branch of phylogenetics is known as phylogeny. Fundamental to phylogeny is the universally accepted belief that all organisms originally descended from a common ancestor (LUCA - last universal common ancestor.)
Evidence for phylogeny comes from various sources. These include analysis of DNA sequences, comparative morphology/anatomy (homology) and palaeontology (analysis of fossils.)
Analysis and comparisons of DNA sequences can involve looking for nucleotide differences in a particular gene. For example, DNA sequences can be examined for any nucleotide mutations in the gene coding for cytochrome-c (a protein found in almost all living things.)
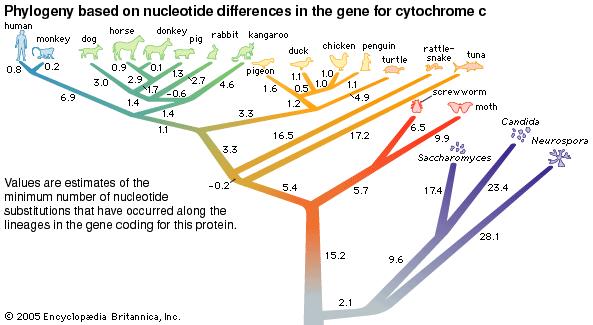
The fewer the nucleotide differences in the sequence, the more closely related the organisms are to each other. One way in which genomic sequences can be observed is through Phylogenetic Footprinting[2]. This is where regulatory sites on DNA are identified, allowing comparative genomics between closely related species; this process is very efficient at identifying DNA sequences controlling gene expression.
The study of phylogenetics is best represented by phylogenetic trees. These effectively present phylogenetic findings in a visual form. Different phylogenetic trees can be integrated in order to produce the most accurate representation of the evolution of modern life forms[3]. The most famous phylogenetic tree is the Universal SSU rRNA Tree of Life. Prior to this new tree, previous trees have stated that there are three domains of cellular life - Bacteria, Archaea and Eukarya - as three sister branches derived from the LUCA. The new generally accepted phylogenetic tree, however, proposes that there were only 2 branches which spring from LUCA - Archaea and Bacteria and that Eukaryotic Domain stems from the Archaeal domain. In order to construct an evolutionary tree from protein sequences, first the genes/proteins being compared need to be aligned in order to be able to make a comparison between the sequences and see how similar they are. These similarities are then converted to evolutionary distance measurements using a mathematical model, so a tree can now be drawn[4].
References
- ↑ Daniel L. Hartland, Elizabeth W. Jones (2009) Genetics: Analysis of Genes and Genomes, 7th edn., Sudbury, Massachusetts: Jones and Bartlett Publishers.
- ↑ Alberts, Johnson, Lewis, Raff, Roberts, Walter (2008) Molecular Biology of the Cell, 5th edn., New York: Garland Science, Taylor and Francis Group.
- ↑ Alberts, Johnson, Lewis, Raff, Roberts, Walter (2008) Molecular Biology of the Cell, 5th edn., New York: Garland Science, Taylor and Francis Group.
- ↑ Professor TM Embley; A taster of molecular phylogenetics and population genetics. Genetics, Newcastle University, 2017
