Converting IDs - Biomart
This is going to seem odd, in fact, it may seem very odd.
However, you have already witnessed this problem in your earlier work on this site.
As you may have noticed, not all datasets (databases) use the same unique identification codes for the same thing. For example, you have been using a lot of Affymetrix IDs such as 218692_at, and you may have spotted that in some of the analyses you have been doing that these Affymetrix IDs map two different IDs in different systems. In fact, if we take 218692_at we will find that it maps to NP_001093226, NP_001093213, NP_001093223, NP_001093217, NP_001093219, NP_001093224, NP_001093221, NP_001093225, NP_001093222, NP_001093218, NP_001093216, NP_001093214 and NP_001093220 in the protein database at NCBI.
In an ideal world, all the different systems with DNA and protein sequences would easily map to the same unique ID. However, sadly this is not the case.
Sometimes in bioinformatics, we have to take IDs from one system and convert them to another so that we can continue our analysis. For example, we may have needed to map the protein ID - NP_001093226 - back to its Affymetrix ID - 218692_at - so we could carry out some analysis. Luckily for us, we don't have to do this by hand as there are a number of systems that can help us.
In this section, we are going to look at one such system which is the Ensembl implementation of a system called Biomart. Yes, the system looks scary but we will walk through it over the rest of the page. It's not that bad. Honest!
You have been carrying out a series of proteomics experiments that have produced a list of proteins that have changed expression levels, when compared to healthy patients, in the pancreas islets cells.
We are now going to look at how we can convert a list of protein IDs (we have used the RefSeq Protein ID (e.g. NP_001005353) identification system) to their equivalent Affymetrix IDs (Affy HG U133A probeset).
- Download the list of protein IDs in this file.
- Go to: http://www.ensembl.org/biomart/martview/

Setting up Biomart takes a bit of work. However, it is fairly straightforward for the type of conversion we are going to do. In fact, the system just needs four pieces of information:
- the database we are going to use
- the dataset we are going to use
- the data which we wish to convert (the system also needs to know the format of the data)
- the type of data we wish to produce
We are now going to set up the system...
At the start the system looks like this:
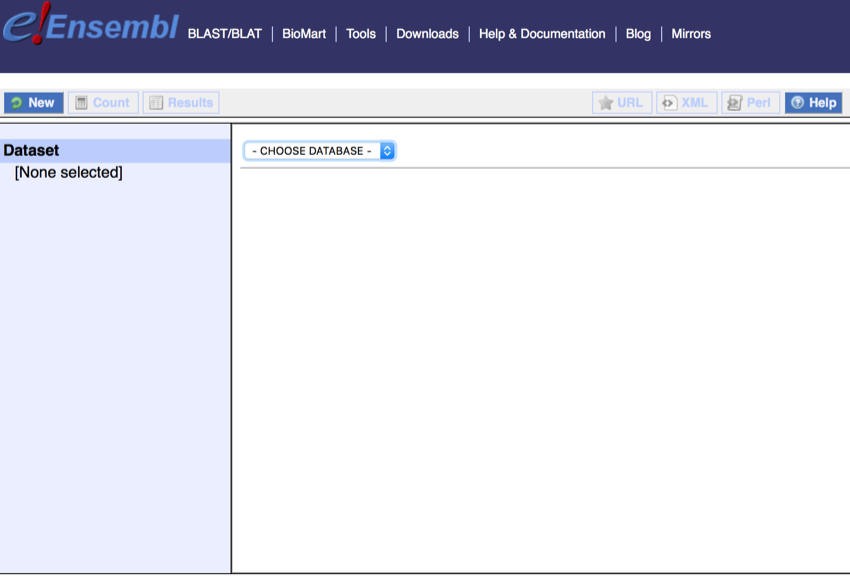
The start screen at Ensembl Biomart
- Change the dataset to 'Ensembl Gene 94'
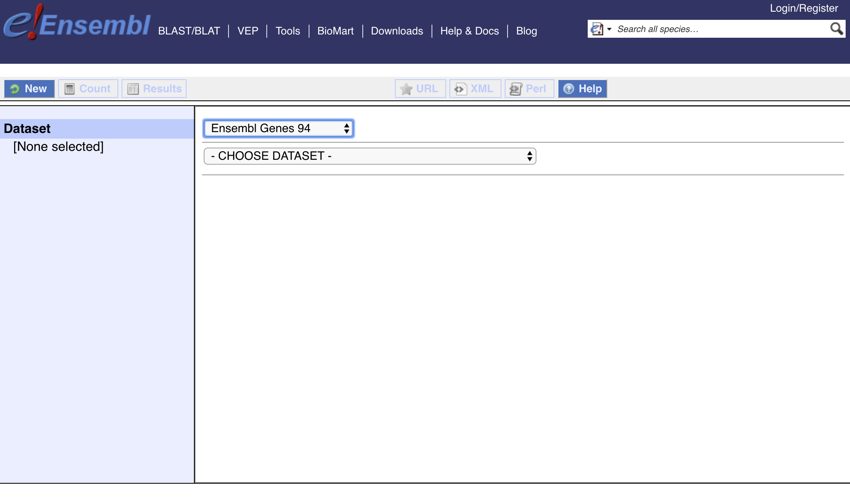
Setting the dataset
- Change the dataset to 'Human genes (GRCh38.p10)'
The screen should now look like this:
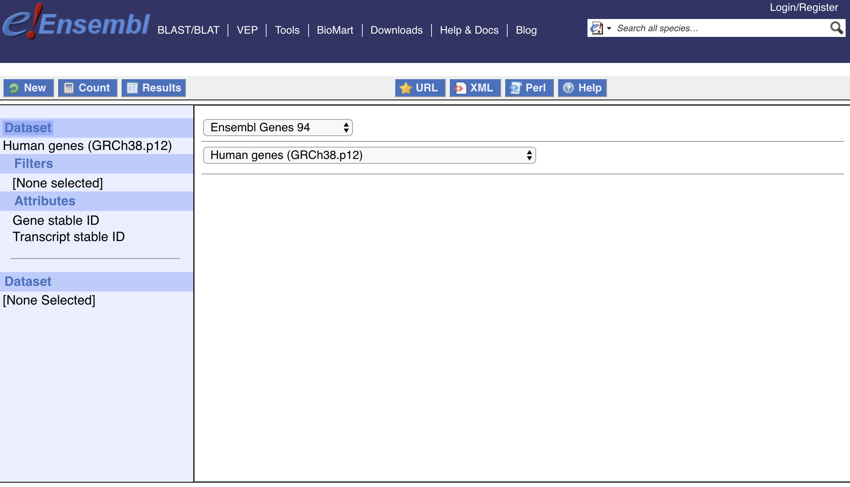
Setting the dataset
Next, we want to set up the 'Filters', this is nothing complicated it's just really asking for the data you wish to convert.
- Click on 'Filters'
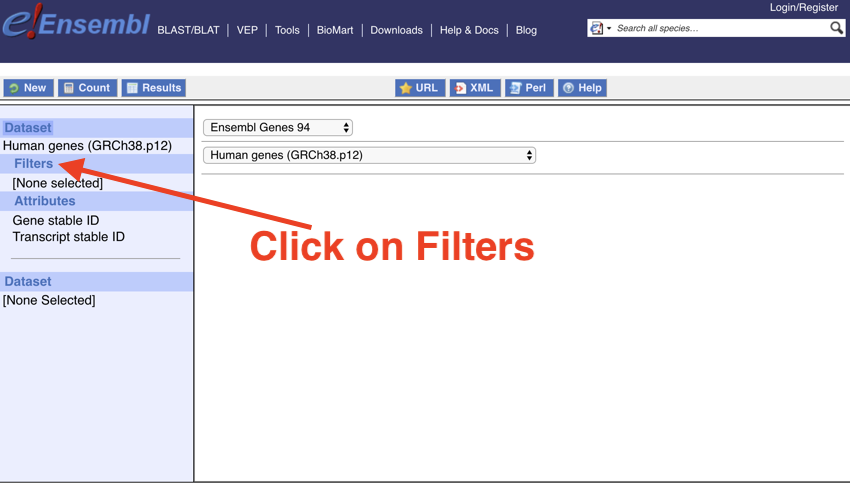
Click on 'Filters'
Your screen should now look like this:
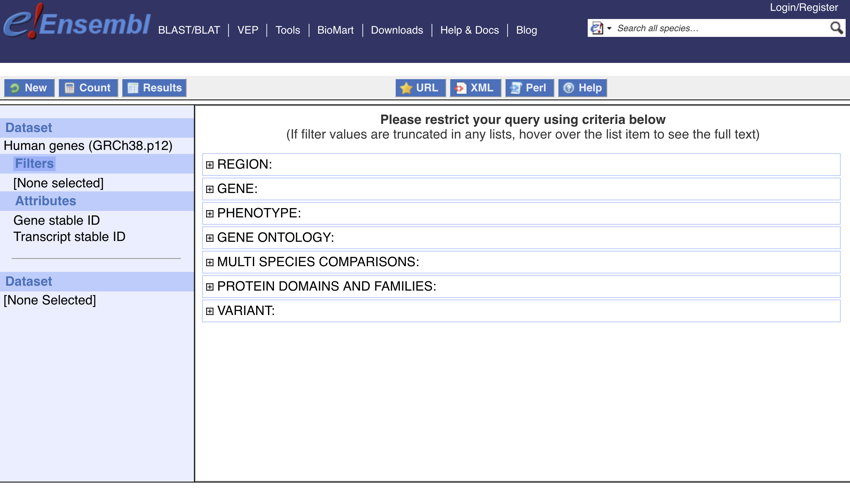
The Filter section
- Click on the + sign next to 'GENE'
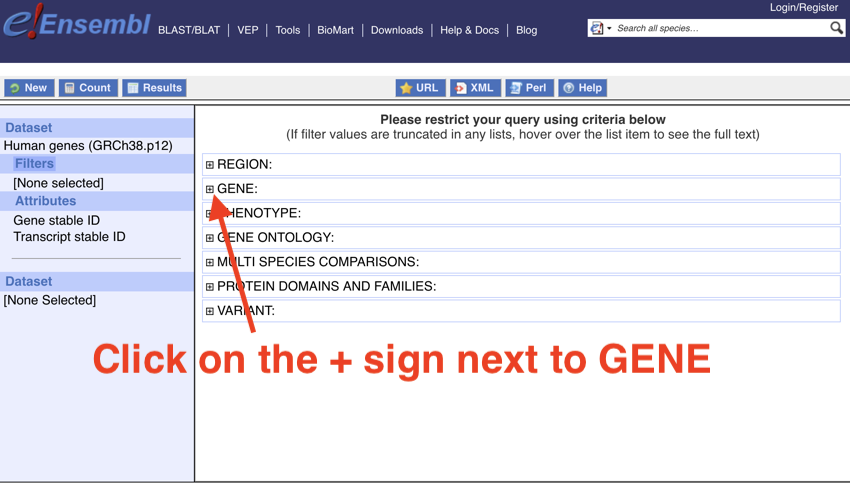
Click on the + sign next to 'GENE'
- On the screen click the checkbox 'Input external references ID list (Max 500 advised)'
- Change the data-type to 'RefSeq Peptide ID(s)' (this may take a bit of scrolling through the menu).
The completed setup should look like the image below.

Setting the filters
- Next, click on 'Choose file'.
- Navigate to the file, and select it.
We have now completed the settings to upload our data. We are now going to set the 'output' (also known as the 'Attributes').
- Click on 'Attributes'.
- Click on 'Gene'.
- Deselect 'Transcript stable ID' and 'Gene stable ID'.
- Click on 'External' (You may need to scroll down).
- Scroll down to 'Microarray probes/probesets (max 2)'
- Click on and select 'Affy HG U133A probeset'.
The 'Attributes' section should now only have one item selected, that is, 'Affy HG U133A probeset'. It should look like the image below:
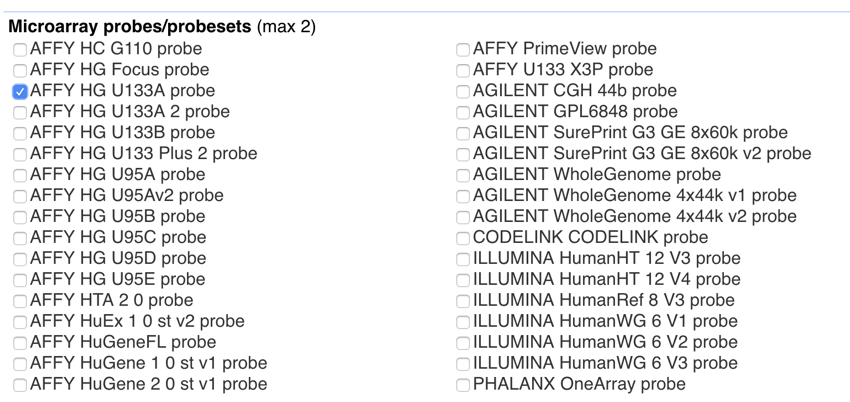
Attributes settings
You have now set up Ensembl Biomart to convert a series of RefSeq Protein ID to there are Affymetrix equivalents.
- Next, click 'Results' (this can be found at the top of the page, top left, ).
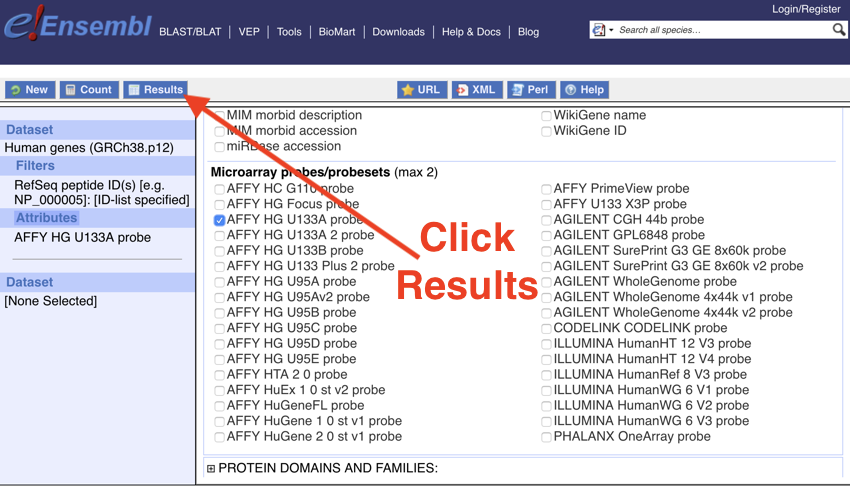
Click the Results
If everything has worked okay then your screen should now look like the image below:
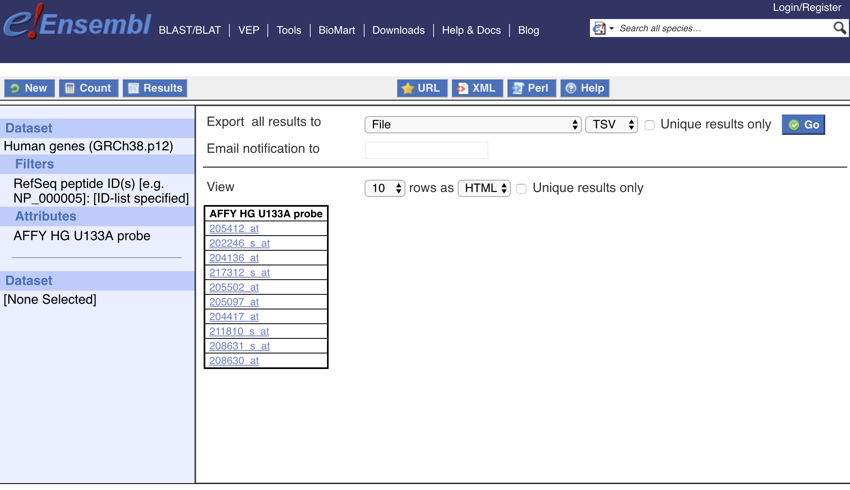
The results
One last thing to do, and that is to download the data.
- Click the 'Go' button. (The download may take some time...)
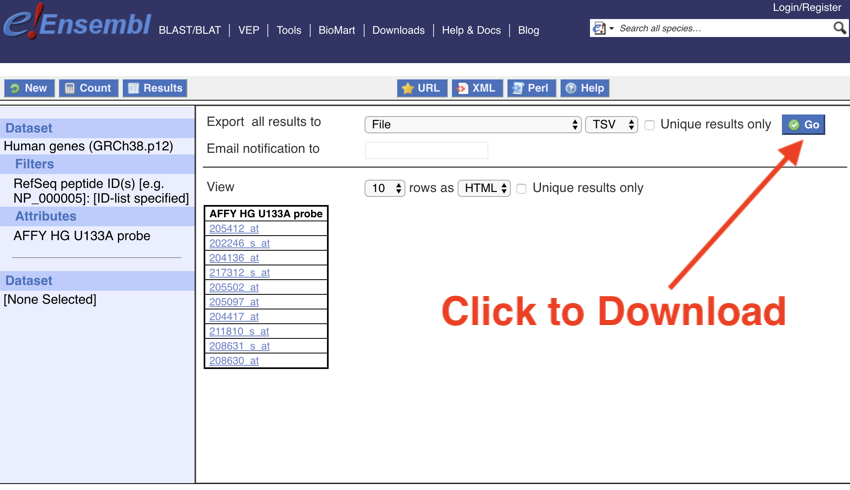
Download the data
You should now have downloaded a file, the contents of which look a bit like the screenshot below:
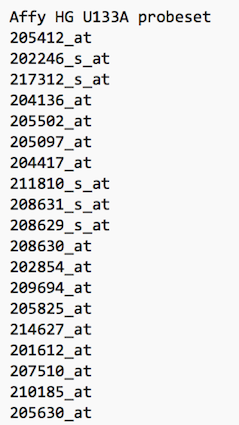
the new dataset
We are now going to use this dataset with DAVID.
- Return to the earlier section of this training site on DAVID and using the dataset you have just downloaded, complete the quiz below.
You have now successfully used DAVID to analyse two different datasets. You also learned how to successfully convert protein IDs to Affymetrix IDs.
The next stop is to use a system called String to find and map relationships between proteins.











